This blog post was originally written by Caroline LaManna, published Mar 8, 2016.
The updated and expanded version by Nyla Naim was published Sept 3, 2020.
Scientists around the world have been making major improvements to CRISPR technology since its initial applications for genome engineering in 2012. Many of these advances have stemmed from the goal of reducing off-target Cas9 activity. The use of nickases, prime editing, anti-CRISPR proteins, and other techniques all aim to improve targeting specificity or reduce the duration of Cas9 activity.
The field of optogenetics is renowned for enabling precise temporal and spatial control. Optogenetics uses genetically encoded tools, such as microbial opsins, to control cellular activities using light. In 2015, scientists combined CRISPR and optogenetic techniques to develop a variety of photoactivatable CRISPR tools. These tools allow scientists to use light to externally control the location, timing, and reversibility of the genome editing process. Read on to learn about the various light-controlled CRISPR tools available to researchers - some readily found at Addgene.

Shining light on transcriptional activation using dCas9
Initial photoactivatable CRISPR systems published in early 2015 focused on using light to control transcription. Two separate labs, Moritoshi Sato’s lab at the University of Tokyo and Charles Gersbach’s lab at Duke University developed similar systems based on the light-inducible heterodimerizing cryptochrome 2 (CRY2) and calcium and integrin-binding protein 1 (CIB1) proteins. The goal of both groups was to create a system that would use light to turn on and off gene expression while imparting spatiotemporal control, reversibility, and repeatability.
The system developed by the Sata lab is composed of two fusion proteins: 1) the genomic anchor - an inactive, dead Cas9 protein (dCas9) fused to CIB1; and 2) the activator - the CRY2 photolyase homology region (CRY2PHR) fused to a transcriptional activator domain (VP64 or p65). Upon expression in the cell, the dCas9-CIB1 fusion binds to the target DNA sequence as directed by the guide RNA (gRNA), while the CRY2PHR-activator fusion floats freely, depicted in the figure below. Once triggered by blue light, the CRY2 and CIB1 proteins heterodimerize and move the activator into position to activate gene transcription. The researchers tested a variety of combinations to optimize both fusion proteins. The best performing combination was NLS-dCas9-trCIB1 and NLSx3-CRYPHR-p65 - it had the lowest background activity in the dark state and highest fold induction at 31X. By using a slit pattern during blue light exposure (470nm), the researchers showed that expression of the human ASCL1 gene could be spatially controlled in a reversible and repeatable manner.
With their light-activated CRISPR/Cas9 effector (LACE) system, the Gersbach lab utilized a similar strategy to develop an optimized photoactivatable CRISPR gene activation system, but settled on a different optimal fusion protein combination. The optimized LACE system consisted of: 1) CIBN-dCas9-CIBN, where CIBN is the N-terminal fragment of CIB1 and it was fused to both the N- and C-termini of dCas9; and 2) CRY2FL-VP64, a fusion of full-length CRY2 and the transcriptional activator domain VP64. Using this system in HEK293T cells to induce expression of human IL1RN, the researchers saw an 11-fold increase in mRNA levels after 2 hr and a 400-fold increase after 30 hr. The system was also shown to be reversible and repeatable when blue light (450nm) was cycled on-off-on. Using a slit photomask, the researchers also demonstrated the ability to spatially control gene expression.
Both of these techniques take advantage of light-induced dimerization to activate their CRISPR system. This common optogenetic technique can be employed in other split systems where light is required to reconstitute enzymatic activity.
Photoactivatable genome modifications by NHEJ and HDR using split systems
Later in 2015, the Sato lab unveiled a photoactivatable system to cleave a target DNA sequence (Nihongaki, et al., 2015) resulting in a double strand break (DSB) that can be repaired by either non-homologous end joining (NHEJ) or homology directed repair (HDR). This system is unique in that it utilizes a “split” nuclease - the authors fragmented Cas9 into N-terminal (residues 2-713, N713) and C-terminal (residues 714-1368, C714) halves, rendering the Cas9 non-functional when split but regaining functionality when the halves are reassociated. By fusing a photoinducible, heterodimerizing domain to each of the Cas9 fragments, the authors created a photoactive Cas9 tool, as shown in the figure below. Their most successful design utilized Magnet photoswitchable proteins derived from the fungal photoreceptor, Vivid (VVD, N. crassa) (Kawano et al., 2015). Nicknamed paCas9-1 and consisting of the fusion proteins N713-pMag and nMagHigh1-C714, this new system had both low background and high fold-induction of Cas9 activity (16.4-fold). This paCas9-1 light-inducible system was able to recognize the same PAM and had similar targeting specificity as full-length Cas9 (flCas9). When triggered by blue light (470nm), paCas9-1 induced indel mutations via NHEJ (frequency of 20.5%) and induced modifications by HDR (frequency of 7.2%).
The authors additionally showed that they could lower the background activity of the system by modifying paCas9-1 using nMagC714 instead of nMagHigh1-C714, generating paCas9-2. This change did not significantly alter the system’s efficiency at generating mutations when activated with light and lowered background DSBs (non detectable). Like their prior work, the Sato lab also showed that the paCas9-2 system could be spatially controlled and reversibly activated by turning blue light on and off.
As one might expect from the modular nature of Cas9, Nihongaki et al. showed that it was possible to swap out the Cas9 domains in their split fusions and generate a photoactivatable nickase and a photoactivatable repressor (dCas9). The activity of all variants was reversible and repeatable.
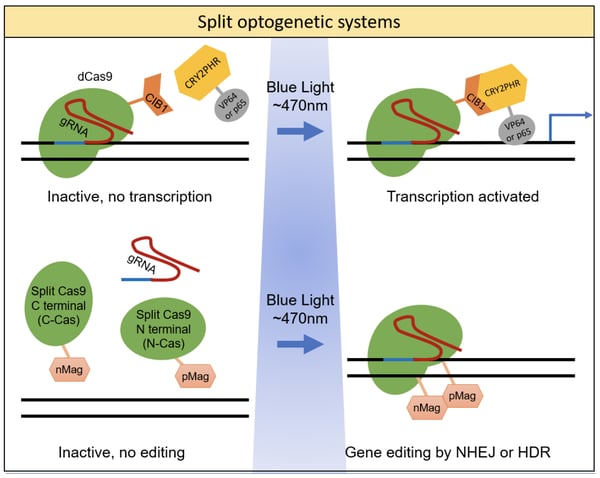
Since these initial studies published in 2015, more split systems have been developed for use with CRISPR-based gene editing (see table below). For example, the far-red light-activated split-Cas9 (FAST) system constitutively expresses the C-terminal fragment of Cas9 while transcription of the N-terminus is light-dependent (Yu et al., 2020). To express the N-Cas9, Haifeng Ye’s lab used the bacterial phytochrome BphS which produces c-di-GMP in response to far-red light (730 nm). This nucleotide triggers the translocation of the protein BldD to the nucleus. To couple this light-dependent activity to transcription, they fused BldD to the transcriptional activators p65 and VP64 which in turn transcribes N-Cas9. The N- and C-Cas9 fragments then dimerize to form a functional protein due to being fused with high-affinity interacting proteins Coh2 and DocS, respectively.
The use of far-red light is of particular interest since blue light has limited applications in vivo. UV and blue light is readily absorbed and scattered by the skin, so it is not able to reach deeper tissues. Additionally, high intensity exposure can cause DNA damage. Far-red light, however, can penetrate 5 mm past the surface of skin and is less phototoxic. Using the FAST system, Ye’s lab could edit a tdTomato fluorescent reporter gene in mice using Minicircle DNA vectors encoding the FAST constructs. This proof-of-concept model opens intriguing possibilities for optogenetic manipulation of CRISPR-based systems in animal models.
Use Anti-CRISPRs to Turn Off Genome Editing
Single-chain Cas9 photoswitches
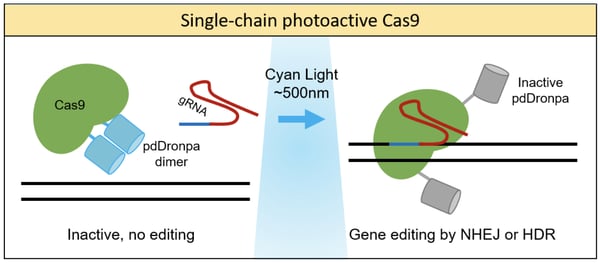
Photoactivation of Cas9 can also be achieved using single-chain fusion proteins by fusing Cas9 to photoreceptors. One prime example was from the lab of Michael Lin, who developed a chimeric protein consisting of Cas9 and a photo-dissociable dimeric green fluorescent protein (pdDronpa) (Zhou et al., 2018). pdDronpa dimers dissociate in response to cyan light (500nm). By fusing the two pdDronpa domains to the sites flanking the DNA-binding cleft of Cas9, the fluorescent domains dimerize and prevent DNA binding in the dark. Upon light stimulation, the pdDronpa moieties move apart, allowing DNA binding and cleavage. This technique was applied to create photoswitchable spCas9, dCas9, and S. aureus Cas9 (saCas9) proteins, showing the versatility of this approach. This study generated the first optically active saCas9 protein.
Stop light! Photoactive anti-CRISPRs halt Cas9 activity
Anti-CRISPR proteins are a highly diverse group of proteins that are able to block CRISPR-Cas systems. They have been used to block genome editing, reduce off-target activity, and prevent cytotoxic effects. For a more detailed review, check out our blog post: Anti-CRISPR Proteins Switch Off CRISPR-Cas Systems.
Listen to the anti-CRISPR podcast segment
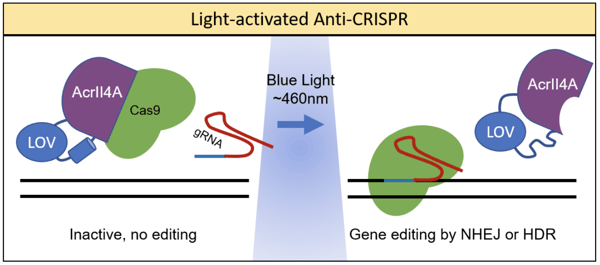
Simply expressing anti-CRISPR proteins does not provide much control over when Cas9 activity is blocked. Hence, co-opting optogenetic techniques allows greater precision and control over Cas9 activity. Dominik Niopek’s lab achieved this by fusing the anti-CRISPR Acr protein, AcrIIA4, to a photo-sensitive LOV2 domain from A. sativa (Bubeck et al., 2018). LOV2 has a C-terminal helix that unfolds when stimulated by blue light. When fused to the catalytic site of AcrIIA4, this unfolding conformational shift can interfere with activity. The resulting system was coined as CRISPR–Cas9 activity switching via a novel optogenetic variant of AcrIIA4 (CASANOVA). As shown in the figure below, in the absence of light, Cas9 is bound by AcrIIA4 and rendered inactive. In the presence of blue light, AcrIIA4 is inhibited by the unfolding of LOV, allowing the release of Cas9.
A noteworthy application of the CASANOVA system was its use in light-mediated CRISPR labeling to study chromatin architecture (Hoffman et al., 2020). Fluorescent dCas9 proteins can be used to identify specific genomic loci for imaging techniques. Since expression of dCas9 can potentially alter the genome and chromatin architecture, it is useful to regulate the time in which dCas9 binding occurs. Bubeck et al. expressed a telomere-targeting gRNA and a dCas9 fusion protein with three red fluorescent tags in U2OS cells. Upon light activation, dCas9 is released within minutes and red foci formed 20-40 min later. The CASANOVA system is versatile in that it can be adapted to regulate gene editing, turn on transcription, label chromatin, and study kinetics.
Using chemistry to photocage CRISPRs
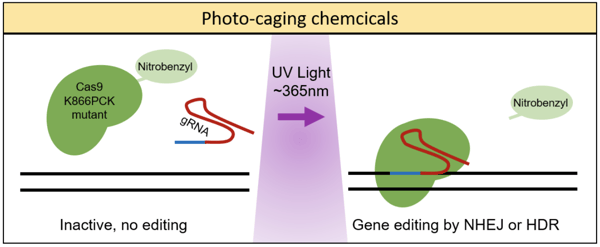
The aforementioned techniques each employed a photoactive strategy which had been engineered from naturally occurring photoactive proteins (i.e. CRY2 and Vivid) - Alexander Deiters’ lab, on the other hand, took a different approach. These researchers used a genetically encoded photocaging technique to insert a light-removable protecting group, specifically a nitrobenzyl photocaged lysine (PCK), on the Cas9 protein (Hemphill et al., 2015). In order to insert the PCK into a specific site on the Cas9, the group used an engineered pyrrolysyl tRNA/tRNA synthetase pair which would add the PCK upon reaching the amber stop codon, TAG. (To learn more about site-specific incorporation of amino acids using pyrrolysl tRNA synthetase, read this article).
The group first tested photocaging various lysines in Cas9 to determine which best deactivated the protein’s ability to cleave targeted DNA, settling on photocaging the K866 lysine, as seen in the figure below. Next, by using a dual reporter fluorescence assay, Hemphill et al. demonstrated that the Cas9 K866PCK mutant was indeed inactive prior to irradiation with UV light (365nm) and that post-UV exposure it showed cleavage activity similar to the wild-type Cas9. This photocaging technique was also shown to impart spatial control of Cas9 cleavage when using a photomasking technique. Last, Hemphill et al. presented data showing that this genetically encoded, photocaged Cas9 system could silence endogenous gene expression - demonstrating light-induced silencing of transferrin receptor CD71 in HeLa cells.
Comparison of photoactivatable CRISPR strategies
| Publication | System nickname | Photoactivatable moiety | Cas9 variatn | CRISPR protein application |
| Split Systems / Light-induced Dimerization | ||||
| Nihongaki et al., 2015 | CRY2 and CIB1 from A. thaliana | dCas9 | Activation of gene transcription | |
| Polstein et al., 2015 | LACE | CRY2 and CIB1 from A. thaliana | dCas9 | Activation of gene transcription |
| Nihongaki et al., 2015 | paCas9 | Magnet proteins from N. crassa | Split Cas9, Split Cas9 nickase, Split dCas9 | Genome editing by NHEJ and HDR, repression of gene expression by CRISPRi |
| Nihongaki et al., 2017 | Split-CPTS2.0 | CRY2 and CIB1 from A. thaliana | dCas9 | Activation of gene transcription |
| Kim et al., 2019 | LADL | CRY2 and CIB1 from A. thaliana | dCas9 | Activation of gene transcription. Label genomic loci. Alter chromatin architecture. |
| Yu et al., 2020 | FAST | BphS from R. sphaeroides | Split Cas9 | Genome editing by NHEJ and HDR. Can be used for in vivo models |
| Single-chain Photoswitchable Cas9 | ||||
| Richter et al., 2016 | tsRC9 | RsLOV from R. sphaeroides | Cas9, dCas9 | Gene editing |
| Zhou et al., 2018 | pdDronpa | Cas9, dCas9 | Gene editing, transcription | |
| Light-Dependent Anti-CRISPR | ||||
| Bubeck et al., 2018 | CASANOVA | LOV2 from A. Sativa | AcrIIA4 anti-CRISPR protein, co-expressed with Cas9 or dCas9 | Repression of gene editing and transcription. Label genomic loci |
| Photochemical Control | ||||
| Hemphill et al., 2015 | Nitrobenzyl-caged lysine | Cas9 K866PCK mutant | Genome editing by NHEJ and HDR | |
| Jain et al., 2016 | CRISPR-plus | Photocleavable ssDNA oligonucleotides | Cas9, photocaged-gRNA | Gene editing |
| Manna et al., 2019 | NPPOC-caged trimethoprim (TMP) | Cas9 fused to destabilizing domains (Cas9-DHFR) | Gene editing, transcription | |
| Zhou et al., 2020 | NPOM-caged nucleobases | Cas9, photocaged-gRNA | Gene editing | |
For more details, check out this comprehensive review comparing optogenetic CRISPR techniques (Matony et al., 2020). Whether you are looking to activate, repress, or modify a gene, you now have the tools at your disposal to control your genome editing using light. We look forward to more tools as CRISPR and optogenetics continue to evolve and can’t wait to see what cool applications you use these for in the future!
References
Bubeck F, Hoffmann MD, Harteveld Z, Aschenbrenner S, Bietz A, Waldhauer MC, Börner K, Fakhiri J, Schmelas C, Dietz L, Grimm D, Correia BE, Eils R, Niopek D (2018) Engineered anti-CRISPR proteins for optogenetic control of CRISPR–Cas9. Nat Methods 15:924–927 . https://doi.org/10.1038/s41592-018-0178-9
Hemphill J, Borchardt EK, Brown K, Asokan A, Deiters A (2015) Optical Control of CRISPR/Cas9 Gene Editing. J Am Chem Soc 137:5642–5645 . https://doi.org/10.1021/ja512664v
Jain PK, Ramanan V, Schepers AG, Dalvie NS, Panda A, Fleming HE, Bhatia SN (2016) Development of Light-Activated CRISPR Using Guide RNAs with Photocleavable Protectors. Angew Chem Int Ed 55:12440–12444 . https://doi.org/10.1002/anie.201606123
Kim JH, Rege M, Valeri J, Dunagin MC, Metzger A, Titus KR, Gilgenast TG, Gong W, Beagan JA, Raj A, Phillips-Cremins JE (2019) LADL: light-activated dynamic looping for endogenous gene expression control. Nat Methods 16:633–639 . https://doi.org/10.1038/s41592-019-0436-5
Mathony J, Hoffmann MD, Niopek D (2020) Optogenetics and CRISPR: A New Relationship Built to Last. In: Methods in Molecular Biology. Springer US, pp 261–281
Manna D, Maji B, Gangopadhyay SA, Cox KJ, Zhou Q, Law BK, Mazitschek R, Choudhary A (2019) A Singular System with Precise Dosing and Spatiotemporal Control of CRISPR‐Cas9. Angew Chem Int Ed 58:6285–6289 . https://doi.org/10.1002/anie.201900788
Nihongaki Y, Furuhata Y, Otabe T, Hasegawa S, Yoshimoto K, Sato M (2017) CRISPR–Cas9-based photoactivatable transcription systems to induce neuronal differentiation. Nat Methods 14:963–966 . https://doi.org/10.1038/nmeth.4430
Nihongaki Y, Kawano F, Nakajima T, Sato M (2015) Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol 33:755–760 . https://doi.org/10.1038/nbt.3245
Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M (2015) CRISPR-Cas9-based Photoactivatable Transcription System. Chemistry & Biology 22:169–174 . https://doi.org/10.1016/j.chembiol.2014.12.011
Polstein LR, Gersbach CA (2015) A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 11:198–200 . https://doi.org/10.1038/nchembio.1753
Richter F, Fonfara I, Bouazza B, Schumacher CH, Bratovič M, Charpentier E, Möglich A (2013) Engineering of temperature- and light-switchable Cas9 variants. Nucleic Acids Research 44: . https://doi.org/10.1093/nar/gkw930
Yu Y, Wu X, Guan N, Shao J, Li H, Chen Y, Ping Y, Li D, Ye H (2020) Engineering a far-red light–activated split-Cas9 system for remote-controlled genome editing of internal organs and tumors. Sci Adv 6:eabb1777 . https://doi.org/10.1126/sciadv.abb1777
Zhou W, Brown W, Bardhan A, Delaney M, Ilk AS, Rauen RR, Kahn SI, Tsang M, Deiters A (2020) Spatiotemporal Control of CRISPR/Cas9 Function in Cells and Zebrafish using Light‐Activated Guide RNA. Angew Chem Int Ed 59:8998–9003 . https://doi.org/10.1002/anie.201914575
Zhou XX, Zou X, Chung HK, Gao Y, Liu Y, Qi LS, Lin MZ (2017) A Single-Chain Photoswitchable CRISPR-Cas9 Architecture for Light-Inducible Gene Editing and Transcription. ACS Chem Biol 13:443–448 . https://doi.org/10.1021/acschembio.7b00603
Additional Resources
- “Optogenetics Meets CRISPR” by Jef Akst on The Scientist
- “Photoactivatable #CRISPR-Cas9 systems!” by Doug Mortlock on A CRISPR Blog
Resources on the Addgene Blog
- Read Other Optogenetics Blog Posts
- Read Our Primer on Optogenetics by Derek P. Simon
- Read Our Interview with Ed Boyden
Resources on the Addgene Website
- Browse Our Collection of Optogenetics Plasmids
- Check out Our Genome Engineering Resources
- Brush up on Your CRISPR Background with Our Guide Pages
Topics: Optogenetics, CRISPR, CRISPR 101, Other CRISPR Tools





Leave a Comment