Originally published May 23, 2017 and last updated Jul 23, 2020 by Jennifer Tsang.
 CRISPR-Cas technology is constantly evolving. Variants of Cas proteins can be used for genome editing, activating gene expression, repressing gene expression, and much more. But there’s one thing that was missing: a way to shut off Cas’s activity. The concern is that the longer Cas remains active in a cell, the greater chances there are for off-target edits to occur. Although methods to switch on Cas activity using light or drugs have been developed, the field lacked an “off-switch” for Cas proteins.
CRISPR-Cas technology is constantly evolving. Variants of Cas proteins can be used for genome editing, activating gene expression, repressing gene expression, and much more. But there’s one thing that was missing: a way to shut off Cas’s activity. The concern is that the longer Cas remains active in a cell, the greater chances there are for off-target edits to occur. Although methods to switch on Cas activity using light or drugs have been developed, the field lacked an “off-switch” for Cas proteins.
Discovery of anti-CRISPR proteins
That is, until the discovery of anti-CRISPR proteins in 2012 (Bondy-Denomy et al., 2012). Anti-CRISPR (Acr) proteins are phage-derived small protein inhibitors of CRISPR-Cas systems that help phages evade the CRISPR-Cas immune system of bacteria. These Acr proteins were originally found in type I CRISPR-Cas systems.
Four years later, the Sontheimer and Davidson labs discovered three anti-CRISPRs for a type II system in Neisseria meningitidis (Nme) (Pawluk et al., 2016). While Streptococcus pyogenes Cas9 (SpyCas9) is the most used and well studied CRISPR system, N. meningitidis can also be used for genome editing. In this study, the labs found that anti-CRISPR prevents Cas9 activity in HEK293 by reducing editing from ~30% in control samples to 0-10% in Acr samples. Acr proteins can also be used to prevent dCas9 tethering to DNA.
Since these early anti-CRISPR studies, scientists have discovered over 50 anti-CRISPR proteins that interact with CRISPR-Cas systems such as Cascade-Cas3, Cas9, Cas12, and Cas13 (Marino et al., 2020). Some Acr proteins are specific to a particular Cas protein while others can inhibit CRISPR enzymes from multiple species of bacteria.
A note on anti-CRISPR nomenclature
Acr family proteins are named for the type of CRISPR-Cas system they block and are numbered in order of discovery (Bondy-Denomy et al., 2018). For example, AcrIIA4 mentioned above indicates that it’s the fourth Acr protein discovered that blocks type II-A CRISPR-Cas systems. A full list of Acr proteins organized by the type of CRISPR-Cas system they block can be found here.
How do anti-CRISPR proteins work?
Anti-CRISPRs proteins are highly diverse but most block CRISPR activity one of three ways:
- Inhibit DNA binding
Example: AcrIIA4 blocks Cas9’s interaction with the PAM site (Dong et al., 2017), and AcrIIC3 causes Cas proteins to dimerize which blocks the PAM recognition site (Harrington et al., 2017). - Inhibit crRNA loading
Example: AcrIIC2 interactions with Cas9 interferes and prevents the correct assembly of the crRNA-Cas complex (Zhu et al., 2019). - Inhibiting DNA cleavage
Example: AcrIIC1 binds to the HNH endonuclease domain of Cas9 and prevents target DNA cleavage (Harrington et al., 2017).
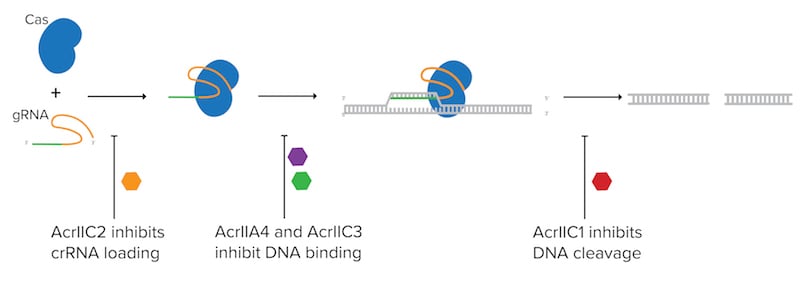 |
| Figure 1: Anti-CRISPR proteins (Acr's) can block CRISPR activity in many ways such as inhibiting crRNA loading, inhibiting DNA binding, or inhibiting DNA cleavage. |
Five ways to anti-CRISPR proteins in your experiments
1. Reduce off-target effects
Prolonged Cas activity can increase the chance of off-target editing particularly after the majority of on-target editing has occurred. Anti-CRISPR proteins can be used to limit this off-target editing, but when is the best time to shut off Cas activity? Using inhibitor timing experiments, the Doudna lab found that at least 50% of on-target Cas9 edits happens within the first six hours. When added to human cells six hours after the introduction of Cas9 RNPs, AcrIIA4 reduced off-target editing while still allowing on-target editing (Shin et al., 2017).
Acrs could also be used to reduce off-target edits of base editors, which convert a nucleotide base pair to another base pair at a specific site.
2. Temporal, spatial, or conditional control of CRISPR activity
Regulating Acr proteins by inducible promoters, light, or small molecules could achieve rapid, dynamic, and spatial control of CRISPR-Cas activity. For example, fusing AcrA4II to the photo-sensitive LOV2 domain allows anti-CRISPR activity to be controlled by blue light (Bubeck et al., 2018).
3. Reduce cytotoxicity of CRISPR-Cas editing in tissues
The small size of Acr proteins (~50-200 amino acids) means they can be delivered in vivo using AAV vectors. Having a way to shut off CRISPR-Cas systems prevents the off-target effects and cytotoxicity that result from excessive nuclease activity in tissues (Li et al., 2018).
4. Selection marker while engineering viruses
Acr genes can be used as positive selectable markers in engineering viruses. For example, an Acr gene was used to replace genes in the difficult-to-engineer Sulfolobus islandicus rod-shaped virus 2 (Mayo-Muñoz et al., 2018). Only viral particles that have the gene deletion could replicate when challenged with the S. islandicus CRISPR-Cas system.
5. Phage therapy
Phage therapy uses phage to treat bacterial infections as an alternative to antibiotics. But because many bacterial pathogens possess CRISPR-Cas systems, phage therapy might not be as effective. This is where Acr proteins come in. Acr proteins are small enough that they could be engineered to “armor” therapeutic phages to protect them from pathogenic bacteria’s CRISPR-Cas system.
Conclusion
CRISPR is a great way to make those genome edits. But, just as important is a way to flip the switch and turn off CRISPR activity. The ability to turn off CRISPR activity is an important step towards CRISPR-based therapies and for more precise editing in the lab.
References and resources
References
Dong D, Guo M, Wang S, Zhu Y, Wang S, Xiong Z, Yang J, Xu Z, Huang Z (2017) Structural basis of CRISPR–SpyCas9 inhibition by an anti-CRISPR protein. Nature 546:436–439 . https://doi.org/10.1038/nature22377
Harrington LB, Doxzen KW, Ma E, Liu J-J, Knott GJ, Edraki A, Garcia B, Amrani N, Chen JS, Cofsky JC, Kranzusch PJ, Sontheimer EJ, Davidson AR, Maxwell KL, Doudna JA (2017) A Broad-Spectrum Inhibitor of CRISPR-Cas9. Cell 170:1224-1233.e15 . https://doi.org/10.1016/j.cell.2017.07.037
Li C, Psatha N, Gil S, Wang H, Papayannopoulou T, Lieber A (2018) HDAd5/35++ Adenovirus Vector Expressing Anti-CRISPR Peptides Decreases CRISPR/Cas9 Toxicity in Human Hematopoietic Stem Cells. Molecular Therapy - Methods & Clinical Development 9:390–401 . https://doi.org/10.1016/j.omtm.2018.04.008
Marino ND, Pinilla-Redondo R, Csörgő B, Bondy-Denomy J (2020) Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat Methods 17:471–479 . https://doi.org/10.1038/s41592-020-0771-6
Mayo-Muñoz D, He F, Jørgensen J, Madsen P, Bhoobalan-Chitty Y, Peng X (2018) Anti-CRISPR-Based and CRISPR-Based Genome Editing of Sulfolobus islandicus Rod-Shaped Virus 2. Viruses 10:695 . https://doi.org/10.3390/v10120695
Pawluk A, Amrani N, Zhang Y, Garcia B, Hidalgo-Reyes Y, Lee J, Edraki A, Shah M, Sontheimer EJ, Maxwell KL, Davidson AR (2016) Naturally Occurring Off-Switches for CRISPR-Cas9. Cell 167:1829-1838.e9 . https://doi.org/10.1016/j.cell.2016.11.017
Shin J, Jiang F, Liu J-J, Bray NL, Rauch BJ, Baik SH, Nogales E, Bondy-Denomy J, Corn JE, Doudna JA (2017) Disabling Cas9 by an anti-CRISPR DNA mimic. Sci Adv 3:e1701620 . https://doi.org/10.1126/sciadv.1701620
Additional Resources on the Addgene Blog
- First time CRISPR user? Check out this post.
- Learn how to Choose the Best Cas9 Variant for Your Next Experiment
- Read more about Optogenetic Control of CRISPR
Resources on Addgene.org
- Find CRISPR Plasmids for Your Research
- Interested in light-inducible Cas9 activity? Check out these these plasmids
- Catch Up on Your CRISPR Background with Our Guide Pages
Other Links
- Check out Pawluk et al's Video Abstract
- Read up on All Things CRISPR with These Papers.
Topics: CRISPR, CRISPR 101






Leave a Comment