This post was contributed by guest blogger Matthew J. Niederhuber, a graduate student at UNC Chapel Hill.
Chromatin immunoprecipitation followed by high-throughput sequencing, ChIP-Seq, is the go-to method for mapping where a protein binds genome-wide, and has been widely applied in many model organisms and cell lines. Although ChIP-seq is a relatively simple and robust protocol it does have limitations. The enzyme-based CUT&RUN method overcomes many of these limitations and makes it easier for you to map protein-DNA interaction with limited biological materials.
Problems with Chip-seq
- ChIP-seq typically requires a large amount of input material, cells or tissue, to produce a strong enough signal over background noise. This is not terribly problematic when working with cell culture, because it is easy to harvest large quantities of cells. However, when working on specific tissues or organs in whole animal models like Drosophila melanogaster, obtaining enough material for a ChIP-seq experiment can be a major technical hurdle.
- ChIP-seq requires cross-linking during an initial fixation step. This serves to stabilize DNA-protein interactions for subsequent sheering of DNA by sonification. Though it helps capture weaker or more transient DNA-protein interactions, the reliance on fixation does increase the chance of false-positives, as proteins may appear to be binding at particular genomic sites when in fact they are just non-specific or indirectly interacting.
The CUT&RUN method
Enter CUT&RUN, an enzyme-based protocol for identifying DNA binding patterns that overcomes some of the limitations of ChIP-seq. CUT&RUN, which stands for ‘Cleavage Under Targets and Release Using Nuclease,’ was recently developed by the Henikoff lab at the University of Washington. CUT&RUN doesn’t require fixation and significantly decreases background noise, reducing the read depth necessary when sequencing and lowering the amount of tissue needed per experiment.
In the McKay lab we study how differential access to transcriptional regulatory elements is controlled during development using the Drosophila wing as a model. A typical ChIP-Seq experiment requires around 1,000 wings. Collecting that much tissue takes a week of multiple people performing dissections. With CUT&RUN, we have had success with significantly fewer wings, such that enough tissue for an experiment can be easily collected in a single day.
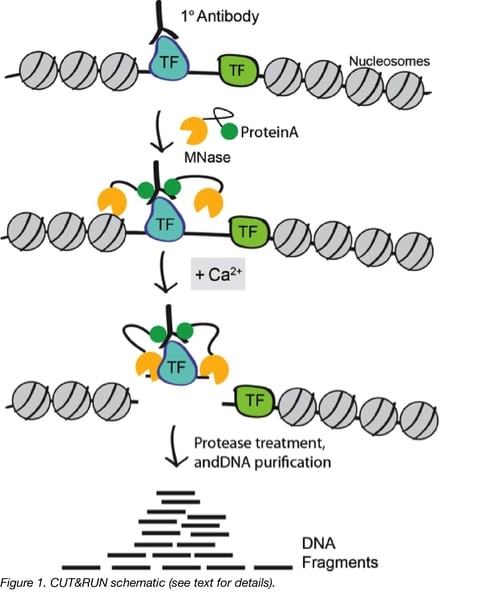
CUT&RUN works by using the DNA cutting activity of a Protein A fused micrococcal nuclease (MNase) to specifically isolate DNA that is bound by a protein of interest. First, nuclei from tissue or cell culture are isolated using lectin-coated magnetic beads. Nuclei are then incubated with a primary antibody against the protein of interest. The Protein A fused MNase is then added and Protein A binds Immunoglobulin G (IgG) thus targeting MNase to antibody bound proteins (Figure 1). Importantly, since Protein A has weaker affinity for mouse IgG, it is helpful to incubate with a rabbit secondary antibody when using a non-rabbit primary, prior to the addition of Protein A MNase.
Once MNase has been localized to target sites, the nuclease is briefly activated to digest the DNA around the target protein. This targeted digestion is controlled by the addition of calcium, which MNase requires for its nuclease activity and is chelated from the reaction up until this point. The nuclease reaction is performed on ice, and only for a short period of time, thus precisely controlling the amount of cutting and mitigating noise generated by off target digestion. After MNase digestion, fragments are released from nuclei by a short incubation at 37 °C. These short DNA fragments can then be purified for subsequent library preparation and high-throughput sequencing.
Improvements to the CUT&RUN method and using it in your own lab
Very recently, the Henikoff lab published a preprint paper outlining improvements to the initial CUT&RUN protocol, the most significant being the usage of a strong detergent to permeabilize cells rather than to rely on nuclei extraction. They report that this change offers a significant improvement and simplification of the original technique, specifically in terms of initial steps of cell or tissue preparation.
All in all, CUT&RUN offers a great alternative to ChIP-seq, especially in situations when it is technically challenging to collect large numbers of cells or when a high degree of signal to noise is desired. It also provides an excellent additional measure of DNA binding sites in combination with existing ChIP-seq data sets, of which there are many readily available. Based on the success that our lab and others have had adopting the CUT&RUN method, it seems likely that this technique will quickly become a standard method alongside ChIP.
Many thanks to our guest blogger, Matthew J. Niederhuber.

Matt Niederhuber is a graduate student at the University of North Carolina in Chapel Hill, where he studies enhancers and blogs about science.
Additional Resources
- Learn how you can use FRET to study protein interactions
- Learn about other types of fusion proteins
- Study chromatin interactions with CAPTURE
Additional Resources on Addgene.org
- Find simple lab protocols
- Browse CRISPR plasmids for isolating a genomic locus
- Browse FRET plasmids for studying protein interactions
Topics: Other Plasmid Tools, Plasmids






Leave a Comment