Originally published Dec 7, 2017 and updated Jul 2, 2020.
Promoters may be the star of gene regulation, but enhancers and chromatin looping play important supporting roles. Enhancers are cis regulatory DNA sequences that, when bound by transcription factors, can increase a gene’s transcription. Sometimes enhancers are located thousands of base pairs away from the gene they regulate, but are brought in proximity by the looping of chromatin, the complex of DNA and proteins that forms chromosomes.
These long range DNA interactions are often detected with chromosome conformation capture (3C) based methods, such as 5C or Hi-C (Han et al., 2018). Think of 3C methods as taking a snapshot of chromatin looping with the photo “developed” by analyzing DNA sequencing data. While “pictures” generated with current 3C methods provide useful information about chromatin interactions, they are grainy so it’s hard to make out the details. To increase the resolution, the Xu lab created a dCas9-based CAPTURE (CRISPR Affinity Purification in situ of Regulatory Elements) method. The original CAPTURE method was published in 2017 and addressed many of the drawbacks of 3C methods, but could only detect chromatin interactions at one location in the genome at a time (Liu et al., 2017). Recently the Xu lab developed CAPTURE 2.0, an updated version of CAPTURE that detects chromatin interactions at hundreds of loci at once (Liu et al., 2020).
CRISPR Affinity Purification in situ of Regulatory Elements: The original CAPTURE method
The original CAPTURE method requires the creation of a stable cell line that co-expresses three components:
- dCas9 with an added biotin acceptor site
- BirA, a biotin ligase
- a gRNA(s) targeting a single genomic location of interest
When these three components are co-expressed, the gRNA targets the dCas9 to the loci of interest and dCas9 is biotinylated by BirA. From here, the CAPTURE method is similar to other 3C methods: crosslinking captures chromatin interactions, a restriction enzyme digest cuts DNA into smaller pieces, and proximity ligation creates chimeric circles of DNA that are distant linearly but close together in space. The chromatin “picture” is then developed by detecting these chimeric DNA fragments by PCR or next-generation sequencing.
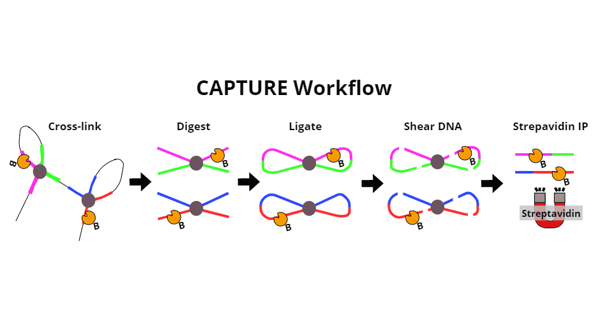 |
| Figure 1: Overview of the key steps of the CAPTURE method. After protein and DNA complexes are cross-linked, DNA is digested into smaller pieces. Sticky ends created are then ligated together to create loops of DNA. This is key to linking pieces of DNA that are far away from each other in terms of base pairs, but close together in terms of where they are physically located. DNA is then sheared into pieces small enough to be pulled down via streptavidin IP. Biotinylated dCas9 goes along for the ride throughout all of these steps, until its connection with DNA is broken during cross-link reversal (not shown in diagram). Cross-links are reversed so that DNA fragments can be detected by PCR or deep sequencing. |
A drawback of the original CAPTURE method is that a new cell line has to be created for each genomic loci of interest. This is cumbersome and also makes it hard to compare results across multiple targets because different CAPTURE cell lines will have varying levels of dCas9 and gRNA expression. Additionally, CAPTURE 3C-based sequencing requires a large number of cells (~5x107). Together, these two requirements prevent the use of CAPTURE with primary cells or rare cell populations.
Key components of the CAPTURE 2.0 system
To improve upon the original CAPTURE method, the Xu Lab made the following changes to create CAPTURE 2.0:
- Replace the biotinalatable tag on dCas9 with a BioTAP-tag. The BioTAP-tag is a 69 aa long biotinylation targeting sequence that is recognized by biotin ligases normally expressed in eukaryotic cells. Using this tag reduces the number of CAPTURE components that need to be delivered to cells from three to two.
- Lentiviral delivery of BioTAP-tagged dCas9 and gRNAs. Using lentiviral delivery of the two remaining CAPTURE 2.0 components eliminates the need to create stable cell lines and allows CAPTURE 2.0 to be used with primary cells.
Find plasmids for CAPTURE 2.0!
When compared to the original CAPTURE, CAPTURE 2.0 had a ~14-fold increase in its capture rate of chromatin interactions at a promoter in the well-characterized beta globin locus. The two methods had similar rates of specificity for the DNA sequence targeted in the experiment.
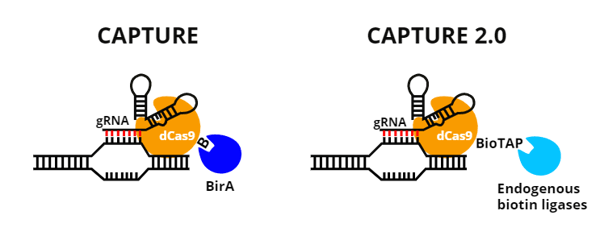 |
| Figure 2: Comparing CAPTURE and CAPTURE 2.0. |
| Original CAPTURE | CAPTURE 2.0 | |
| Biotinylation enzyme | BirA | Endogenous biotin ligases |
| Expression system | Stable cell line expressing 1) FLAG-biotin-tagged dCas9, 2) BirA, and 3) one or more gRNAs | Two separate lentiviruses delivering 1) a BioTAP-tagged dCas9 and an eGFP tag, and 2) gRNAs |
| Number of loci targeted | One | Many |
| Sample type(s) | Cell lines | Cell lines, primary cells |
Table 1: Key differences between original CAPTURE and CAPTURE 2.0 methods.
Applications of CAPTURE 2.0
CAPTURE 2.0’s increased rate of detecting chromatin interactions opens the door for answering more questions about chromatin conformations. The Xu lab presented three ways to use CAPTURE 2.0:
1. Multiplex capture of chromatin interactions
The Xu lab performed a proof-of-concept experiment to show that CAPTURE 2.0 could detect multiple chromatin interactions at once. Chromatin interactions at five sites in the well-characterized beta-globin locus were analyzed with CAPTURE 2.0, with two gRNAs targeting each site. They found that DNA sequences targeted by the gRNAs were the top enriched sequences in the experiment and there was little enrichment of DNA targeted by a negative control gRNA or predicted gRNA off-target sites. Long-range chromatin interactions identified with the multiplexed CAPTURE 2.0 largely replicated interactions previously identified with the original CAPTURE method. Both CAPTURE methods outperformed other 3D chromatin methods such as ChIA-PET or Hi-C, by increasing both the number of unique chromatin interactions identified and the level of on-target enrichment.
2. Determining the spatial and hierarchical organization of chromatin interaction networks
A second application for CAPTURE 2.0 is the construction of spatial and hierarchical chromatin interaction networks. Think of these chromatin interactions networks as a social network. They can tell you things like which enhancers are interacting with which promoters, what part of a gene enhancers like to interact with, and how many promoters or genes each enhancer is interacting with. Super enhancers are particularly good models for building chromatin interaction networks since they are a large cluster of enhancers. CAPTURE 2.0’s multiplexing and high resolution allowed the Xu Lab to simultaneously characterize the ‘social networks’ of 157 super enhancers.
3. Measuring temporal changes in chromatin interactions
A third application for CAPTURE 2.0 is detection of temporal changes in chromatin conformation, like those that occur during development and cellular differentiation. Using a cell line model, the Xu Lab tracked enhancer-promoter interactions during erythroid differentiation.
CAPTURE 2.0 captures chromatin's dynamic role in gene regulation
The high resolution and multiplexed chromatin ‘photos’ generated with CAPTURE 2.0 allow us to uncover new details and patterns in chromatin interactions. The chromatin story told by CAPTURE 2.0 is important, but it’s not the only plot line in gene regulation. When chromatin conformation data is compared with changes in gene expression, chromatin accessibility, and epigenetic modifications, we can start to build a timeline of events that surround changes in gene expression and determine who’s a key driver of these changes. By generating a more vivid chromatin picture, CAPTURE 2.0 re-writes the script on gene regulation.
References
Han, J., Zhang, Z., & Wang, K. (2018). 3C and 3C-based techniques: the powerful tools for spatial genome organization deciphering. Molecular Cytogenetics, 11. https://doi.org/10.1186/s13039-018-0368-2
Liu X, Zhang Y, Chen Y, Li M, Zhou F, Li K, Cao H, Ni M, Liu Y, Gu Z, Dickerson KE, Xie S, Hon GC, Xuan Z, Zhang MQ, Shao Z, Xu J (2017) In Situ Capture of Chromatin Interactions by Biotinylated dCas9. Cell 170:1028–1043.e19 . https://doi.org/10.1016/j.cell.2017.08.003
Liu, X., Chen, Y., Zhang, Y., Liu, Y., Liu, N., Botten, G.A., Cao, H., Orkin, S.H., Zhang, M.Q., & Xu, J. (2020). Multiplexed capture of spatial configuration and temporal dynamics of locus-specific 3D chromatin by biotinylated dCas9. Genome Biology, 21. https://doi.org/10.1186/s13059-020-01973-w
Additional Resources on the Addgene Blog
- First time CRISPR user? Check out our tips for 1st time CRISPR users
- Learn how to Choose the Best Cas9 Variant for Your Next Experiment
- Learn More About Designing gRNAs Against Your Favorite CREs Here
Additional Resources on Addgene.org
- Find Other Cas9 Variants for Purifying Genomic Regions of Interest Here
- Brush Up on Your CRISPR Background with Addgene’s CRISPR Guide Page
Topics: CRISPR, Other CRISPR Tools






Leave a Comment