This post was contributed by Kusumika (Kushi) Mukherjee, a Postdoc at Massachussetts General Hospital.
A little over a decade back when Yamanaka and colleagues reported that it is possible to reprogram differentiated cells into induced pluripotent stem cells (iPSCs) by the addition of reprogramming (or “Yamanaka”) factors, they changed the landscape of regenerative medicine. Their work opened up vast possibilities for the clinical and therapeutic applications of iPSCs. The generation of human iPSCs (hiPSCs) now provides an opportunity to develop and use patient-specific somatic cells that are otherwise difficult to obtain. These can then be used to perform cell therapy and to model diseases in vitro.
Find stem cell plasmids at Addgene
Many sources of adult somatic cells have been probed for their suitability in iPSC programming spanning from fibroblasts and keratinocytes to peripheral blood mononuclear cells (PBMCs). PBMCs isolated from whole blood present many benefits for the production of iPSCs:
- Blood is easily and routinely drawn from patients in contrast to the process of collecting fibroblasts, which require a more invasive skin biopsy procedure. This becomes a major benefit when recruiting patients, specifically children and patients with skin disorders for research and clinical trials, as it allows access to a larger number of willing participants.
- While fibroblasts obtained from aged patients are reprogrammed less efficiently, no such correlation has been found between the age of the patients from whom the blood is obtained and the subsequent reprogramming success [1].
- Fibroblasts need to be cultured and expanded in vitro for several passages to produce an adequate number of starting cells for reprogramming while PBMCs collected from blood are reprogrammed immediately after collection [2].
- PBMCs display unique epigenetic signatures that are closer to hiPSCs than age-matched fibroblasts [3].
These advantages make PBMCs the ideal source of somatic cells for reprogramming purposes. In this post, I’ll describe an episomal vector system that increases reprogramming efficiency through the expression of a shRNA against p53, a modified reprogramming factor cocktail, and viral protein EBNA-1.
Methods for reprogramming hiPSCs from PBMCs
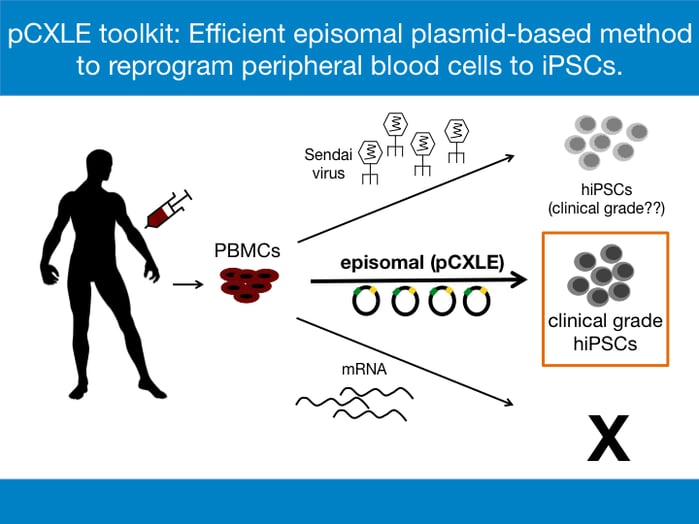
While the method of choice for reprogramming often depends on the source of the somatic cells, the common goal of all reprogramming methods is the expression of the reprogramming factors- OCT3/4, SOX2, KLF4, and C-MYC [1]. For the derivation of clinical grade hiPSCs, reprogramming methods that do not involve modification of the original genome by integrating viral vectors are preferred. This is to avoid the chances of insertional mutagenesis, which is the integration of transgenes into the genome that can cause disruption of functional genes at the site of insertion and which can potentially promote tumorigenesis in vivo. The phenomenon has been recorded in past gene therapy trials and remains a safety concern in the clinical field [4]. Three major reprogramming methods are considered when generating hiPSCs for clinical purposes:
- mRNA transfection- this allows transient expression of the reprogramming factors
- Sendai virus- a non-integrating RNA virus
- Episomal vectors- extrachromosomally maintained DNA plasmids that express reprogramming factors
To date, the mRNA transfection method has not been used successfully to derive iPSCs from PBMCs. Most laboratories use Sendai virus to reprogram PBMCs efficiently [5]. However, the Sendai virus method is expensive, especially if used to make clinical grade hiPSCs. For example, the CytoTune™-iPS 2.1 Sendai Reprogramming Kit designed to generate clinical grade hiPSCs is sold at $11,000 at ThermoFisher Scientific and can be used for only 6-8 reprogramming reactions. Another drawback of using the Sendai virus is the persistence of residual viral material [6]. The complete removal of the residual viral particles and establishment of virus-free hiPSCs usually require about 5-10 passages. Furthermore, working with Sendai virus requires stringent biosafety measures, thereby increasing the costs of the procedure. Episomal reprogramming is an ideal and cost-effective method for creating clinical-grade, safe, nonviral and nonintegrating hiPSCs where the vectors encoding reprogramming factors behave as separate extrachromosomal elements in the nucleus of the target cell. The method also reduces biosafety concerns involved in production and usage of Sendai virus.
Reprogramming efficiency of PBMCs with episomal vectors
Episomal vectors are based on the Epstein-Barr Nuclear Antigen-1 (oriP-EBNA-1) system where each vector contains a viral origin of DNA replication, OriP and the EBNA-1 sequence that encodes for the DNA binding protein EBNA-1 [7]. These two sequences are necessary and sufficient for retention and replication of the vectors in human cells [8]. The EBNA-1 protein recognizes OriP and induces amplification of the episomes concurrently with DNA amplification in the host cell. This enables relatively high and long-term expression of the reprogramming factors. However, reprogramming of PBMCs by episomal vectors has yielded hiPSCs at a low efficiency (0.001-0.03%) [3, 9]. In 2011, Chou et al. generated a mixture of two vectors that encoded six factors - OCT3/4, SOX2, KLF4, C-MYC, LIN28 and SV40 large T antigen (SV40LT). The method had a reprogramming efficiency of only 0.001% when using PBMCs as the source of somatic cells. This translates to only about 16 hiPSC colonies per 100 ml of starting blood sample, in the best-case scenario [7]. In the clinical setting where patient blood samples are limited, such low efficiency of reprogramming is a major hindrance in using PBMCs as a source of somatic cells for reprogramming.
Enhancing PBMC reprogramming efficiency: the pCXLE episomal vector toolkit
It has been shown previously that suppression of TP53 enhances iPSC generation as does replacing C-MYC with L-MYC [10, 11]. While the mechanisms underlying the enhancement of iPSC generation in the presence of these two factors are not completely understood, the addition of an shRNA for P53 and replacing C-MYC with L-MYC in the episomal reprogramming cocktail by Okita et al. increased the reprogramming efficiency in PBMCs [7]. The pCXLE episomal vector system generated by the group is an optimized mixture of three episomal plasmids (Table 1) encoding the reprogramming factors. The episomal vectors are introduced into cells by electroporation. hiPSC colonies are obtained by 10 days after electroporation, most of which are integration-free, karyotypically normal and are able to differentiate into various cell types of all three germ layers [7].
To further enhance the reprogramming efficiency, an extra plasmid, pCXWB-EBNA-1 is used in the reprogramming cocktail. pCXWB-EBNA-1 lacks oriP and is unable to replicate in human cells, thus expressing additional EBNA-1 transiently. The transiently expressed EBNA-1 increases protein expression from the other episomal plasmids. When used on PBMCs, the pCXLE episomal vector system increases the efficiency of reprogramming from 0.001% to nearly 0.1% [7]. In terms of hiPSC colonies, nearly 16 hiPSC colonies can now be obtained from just 1 ml of peripheral blood, a huge improvement from the previously reported efforts. The pCXLE episomal system thus provides a practical way to generate transgene-free, virus-free clinical grade hiPSCs from easily accessible patient blood samples. All pCXLE vectors listed below in Table 1 are available at Addgene.
Table 1: List of pCXLE episomal plasmids, their Addgene ID numbers and the reprogramming factors they encode
| Episomal Plasmid | Addgene ID# | Encodes |
| pCXLE-hSK | 27078 | SOX2 and KLF4 |
| pCXLE-hUL | 27080 | L-MYC and LIN28 |
| pCXLE-hOCT3/4-shp53-F | 27077 | OCT3/4 and shRNA against p53 |
| pCXWB-EBNA1 | 37624 | transient EBNA-1 |
Many thanks to our guest blogger, Kusumika (Kushi) Mukherjee.
 Kusumika (Kushi) Mukherjee is currently a postdoctoral fellow at Massachusetts General Hospital with research interests in the fields of gene editing and stem cells. Connect with her on LinkedIn @ https://www.linkedin.com/in/
Kusumika (Kushi) Mukherjee is currently a postdoctoral fellow at Massachusetts General Hospital with research interests in the fields of gene editing and stem cells. Connect with her on LinkedIn @ https://www.linkedin.com/in/
References
1. El Hokayem, Jimmy, Holly N. Cukier, and Derek M. Dykxhoorn. "Blood Derived Induced Pluripotent Stem Cells (iPSCs): Benefits, Challenges and the Road Ahead." Journal of Alzheimer's disease & Parkinsonism 6.5 (2016). PubMed PMID: 27882265. PubMed Central PMCID: PMC5118044.
2. Kim, Y., et al., The Generation of Human Induced Pluripotent Stem Cells from Blood Cells: An Efficient Protocol Using Serial Plating of Reprogrammed Cells by Centrifugation. Stem Cells Int, 2016. 2016: p. 1329459. PubMed PMID: 27579041. PubMed Central PMCID: PMC4989082.
3. Chou, B.K., et al., Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res, 2011. 21(3): p. 518-29. PubMed PMID: 21243013. PubMed Central PMCID: PMC3193421.
4. Hacein-Bey-Abina, S., et al., LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science, 2003. 302(5644): p. 415-9. PubMed PMID: 14564000.
5. Seki, T., S. Yuasa, and K. Fukuda, Generation of induced pluripotent stem cells from a small amount of human peripheral blood using a combination of activated T cells and Sendai virus. Nat Protoc, 2012. 7(4): p. 718-28. PubMed PMID: 22422317.
6. Diecke, S., et al., Recent technological updates and clinical applications of induced pluripotent stem cells. Korean J Intern Med, 2014. 29(5): p. 547-57. PubMed PMID: 25228828.
7. Okita, K., et al., An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells, 2013. 31(3): p. 458-66. PubMed PMID: 23193063.
8. Ehrhardt, A., et al., Episomal vectors for gene therapy. Curr Gene Ther, 2008. 8(3): p. 147-61. PubMed PMID: 18537590.
9. Mack, A.A., et al., Generation of induced pluripotent stem cells from CD34+ cells across blood drawn from multiple donors with non-integrating episomal vectors. PLoS One, 2011. 6(11): p. e27956. PubMed PMID: 22132178. PubMed Central PMCID: PMC3222670.
10. Hong, H., et al., Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature, 2009. 460(7259): p. 1132-5. PubMed PMID: 19668191. PubMed Central PMCID: PMC2917235.
11. Nakagawa, M., et al., Promotion of direct reprogramming by transformation-deficient Myc. Proc Natl Acad Sci U S A, 2010. 107(32): p. 14152-7. PubMed PMID: 20660764. PubMed Central PMCID: PMC2922531.
Additional Resources on the Addgene Blog
- Stem Cell Models for Disease & Open Science: Interview with Darrell Kotton
- Four Factors that Differentiate the Stem Cell Field
- Sign Up to Write for the Addgene Blog!
Resources on Addgene.org
- Browse Our Stem Cell Collection
- Transcription Factor Fusions for Enhanced Reprogramming
- Find plasmids from the Yamanaka Lab
Topics: Stem Cells, Other Plasmid Tools, Plasmids






Leave a Comment