 This article is based on an interview with Novartis researcher, Carrie Bhang.
This article is based on an interview with Novartis researcher, Carrie Bhang.
The ClonTracer Library, deposited by Carrie Bhang, a research investigator in the In Vivo Pharmacology group at Novartis Oncology, is an exciting new tool that allows researchers to individually label millions of mammalian cells through lentiviral infection and to monitor their abundance and clonal dynamics over time using next generation sequencing (NGS). The library was developed when Carrie was a post-doc in Frank Stegmeier’s lab in Novartis Oncology.
Bhang did her PhD work at Johns Hopkins and came to Novartis for her post-doc in January 2012. She worked for three years as a post-doc in the Target ID and Validation Group and says that she decided to do a post-doc at Novartis because she wanted first-hand experience in industry research. When she came to Novartis, she was happily surprised that “Novartis was actually really research oriented [and] somewhat similar to an academic research environment” with a strong focus on publishing, but still made it so she could “get exposure to the drug discovery process.”
The ClonTracer library originally came about as Bhang and co-workers were trying to develop a way to quantify the take rate of mouse xenografts after cell implantation. Their goal was to figure out how to optimize protocols for in vivo screening with lentiviral pooled shRNA libraries. Later, Bhang says, “When we developed this tool, there were so many interesting questions that we could apply [it to].”
Using ClonTracer to study drug resistance
Although Bhang and co-workers started using the tool to look at a variety of questions surrounding cancer development, metastasis, and drug resistance, Bhang says “the [drug] resistance story panned out quickly,” and was summarized in a recent Nature Medicine paper. In the publication, Bhang and coworkers used the ClonTracer library to determine whether or not cancer cell populations resistant to drug treatments arise during treatment or are preexisting in the parent population prior to the start of treatment. They hypothesized that, if they labeled the parent cell population using the library, propagated it, separated it into roughly identical replicate groups, and treated these groups with the same drug regimen, they would find one of two outcomes (see schematic depiction below):
1. Resistant cells among the separate replicates would contain the same labels - i.e. they would always come from the same parent cells that represent the preexisting resistant subpopulation
2. Resistant cells among the separate replicates would have different labels - i.e. they would stochastically arise from different cells throughout the treatment regimen and were not preexisting.
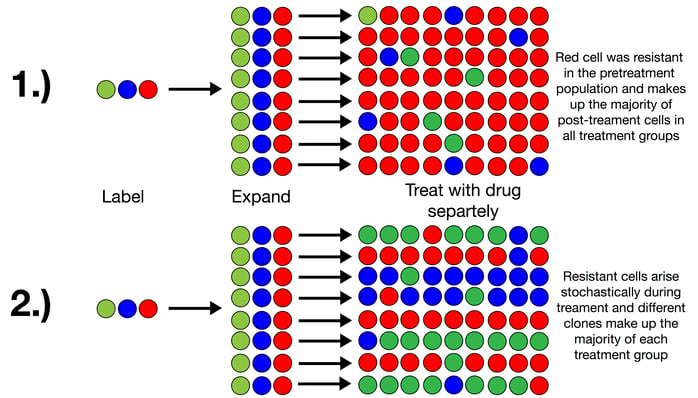
This was previously debated in the cancer field and was difficult to determine using NGS because the error rate of current NGS approaches allows for a detection limit of roughly 0.1%. Thus, rare, pre-existing clones that are present at less than 0.1% of the total population are easily be missed.
The ClonTracer library allows a researcher to label individual members of a starting population of cells with specifc, easily read sequences that can be used to measure their clonal abundance before and after a treatment. The library consists of a pool of lentiviral vectors with roughly 73 million semi-random, 30 bp DNA barcodes that are integrated into target cells' genomes upon infection. When using the ClonTracer system, lentiviruses containing these barcodes infect the target cell population at low multiplicity of infection (MOI) such that each individual cell receives only one barcode. The abundance of each barcoded clone can then be monitored over time by sequencing the barcodes in the population (all barcodes can be amplified using the same sets of forward and reverse primers). The abundance of a particular barcode sequence then corresponds to the abundance of a specific clone; lineage tracing is performed via barcode sequencing.
Using the ClonTracer library, Bhang et al. found that antibiotic resistant cancer cells exist prior to drug treatment. They infected a non-small cell lung cancer cell line containing an activating mutation in epidermal growth factor receptor (EGFR) with the ClonTracer library and monitored population dynamics after treatment with EGFR inhibitor, erlotinib. Looking at barcode abundances among multiple replicate populations, they found that similar barcodes consistently made up the majority of the resistant populations. In fact, 88% of the barcodes within the resistant populations were shared by at least one other replicate; prior to treatment, there are small (previously undetectable) populations of cancer cells that are resistant to drug treatment and that these are selected for during the treatment process. The authors went on to show that combination treatment with multiple drugs reveals even smaller preexisting populations with resistance to both drugs.
The future of ClonTracer
Since starting her new role as an investigator in the In Vivo Pharmacology group at Novartis Oncology, Bhang has been working on using the ClonTracer library to monitor clonal dynamics following drug treatment in cell line and primary tumor xenograft models. She hopes that the library can be used to identify appropriate, non-overlapping targets for drug development - such that initial resistant populations can be eradicated up-front quickly. This approach is in contrast to the current treatment paradigm where resistant populations are treated as they arise. In their Nature Medicine paper, Bhang and coworkers also looked at cancer cell resistance to ABL1 inhibitors in chronic myeloid leukemia. Bhang says the work highlighted the importance of developing more potent ABL1 allosteric inhibitors at Novartis.
The ClonTracer library has been distributed all over the world and was recently used in a study focusing on tumor cell resistance to EGFR inhibitor (Hata et al., 2016). Bhang deposited the library with Addgene because she wanted to make it as easy as possible for researchers to use this tool in their own work. She says, “I spend a lot of time packing, shipping, and sharing protocols one by one,” and she is excited to deposit the library with Addgene because it is a “really great way of broadly sharing these kinds of reagents.”
Here at Addgene, we’re excited to see the creative new ways you put the ClonTracer library to use!
Many thanks to Carrie and Novartis for taking the time to talk with us and for providing this powerful resource to the Addgene community!
 Carrie Bhang is a research investigator in In Vivo Pharmacology at Novartis Oncology and is particularly interested in applying what we have learned about tumor heterogeneity to drug discovery and the development of combination therapies.
Carrie Bhang is a research investigator in In Vivo Pharmacology at Novartis Oncology and is particularly interested in applying what we have learned about tumor heterogeneity to drug discovery and the development of combination therapies.
References
1. Hyo-eun, C. Bhang, et al. "Studying clonal dynamics in response to cancer therapy using high-complexity barcoding." Nature medicine 21.5 (2015): 440-448. Pubmed PMID: 25849130.
2. Hata, Aaron N., et al. "Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition." Nature medicine (2016). PubMed PMID: 26828195. PubMed Central PMCID: PMC4900892.
Additional Resources on the Addgene Blog
- Learn about the Sabatini and Wood Drug Screening Kit
- Find Out How to Best Utilize Our Cancer Resource Pages
Resources at Addgene
- Find the ClonTracer Library at Addgene
- Find Lentiviral Plasmids at Addgene
- Check out Our Cancer Resources Page
Topics: Viral Vectors, Cancer, Cell Tracing, Retroviral and Lentiviral Vectors






Leave a Comment