 Cancer – a term familiar to almost every person regardless of profession, race, or gender. Almost no other disease receives as many billions of dollars in funding or as much attention collectively from researchers. Scientists, however, have yet to slay the beast. On a global scale, tens of millions of new cancer cases arise and millions of people die from cancer-related complications each year. Despite these grim statistics, new strategies for attacking cancer are yielding promising results in the laboratory, and Addgene is accelerating this research by helping to make new cancer resources easily accessible to the research community.
Cancer – a term familiar to almost every person regardless of profession, race, or gender. Almost no other disease receives as many billions of dollars in funding or as much attention collectively from researchers. Scientists, however, have yet to slay the beast. On a global scale, tens of millions of new cancer cases arise and millions of people die from cancer-related complications each year. Despite these grim statistics, new strategies for attacking cancer are yielding promising results in the laboratory, and Addgene is accelerating this research by helping to make new cancer resources easily accessible to the research community.
What is cancer, really?
While cancer is extraordinarily complex at the molecular level, at the cellular level there is a universal theme - uncontrolled cellular proliferation. We have appreciated for some time the causes of this uncontrolled proliferation, in particular the roles of proto-oncogenes, tumor suppressors, and their protein products. Proto-oncogenes consist of a group of genes/proteins that, when expressed, normally encourage cell survival, growth and proliferation, while tumor suppressors perform the opposite task, keeping cells from dividing out of control. One may compare the roles of proto-oncogenes and tumor suppressors to the accelerator and brake pedal of a vehicle, respectively. In the case of a mutation that turns a proto-oncogene permanently “on”, (now an oncogene), this is analogous to the accelerator of a vehicle being stuck down. Similarly, a mutation that turns a tumor suppressor “off” is analogous to a brake pedal that does not work. Both cases yield the same result – uncontrolled movement of the vehicle or, in the case of cancer, uncontrolled cell proliferation. Hence it is imperative to the health of an organism that both classes of genes function normally to maintain balance – encouraging cells to grow and divide only at the right times and in the right places.
Checks and balances: protecting against cancerous cells
Luckily, there are several safety mechanisms in place to protect us from rogue cells when something does go awry. The process of programmed cell death, or apoptosis, is one such mechanism. When a cell or its genetic material is damaged beyond repair, that cell will most often commit itself to the apoptotic pathway and be eliminated from the body. In the case of cancer, however, the apoptotic pathway is circumvented because of the specific genes involved. Downregulation of tumor suppressors and upregulation of proto-oncogenes can directly inhibit the ability of a cell to undergo programmed cell death while simultaneously stimulating growth and division. The combined effect is unregulated proliferation.
Another safety net is our own immune system, which can recognize antigens on cancerous cells and eliminate them from the body. While effective, the immune system does not always eliminate every cancerous cell. Even so, to pose the greatest threat, cancerous cells still need to successfully infiltrate the circulatory or lymph systems to spread, or metastasize. Many cancerous cells will die trying, as the circulatory and lymph systems are not ideal environments for many cell types.
From proteins to pathways - Ras as a model
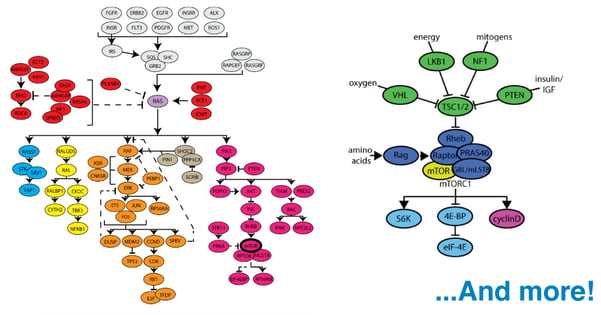
When mutations do arise and cancer ensues, traditional chemotherapies have taken advantage of the universally-dividing behavior of cancer cells, attacking the cellular machinery necessary for division. This approach also kills dividing non-cancerous cells, such as those within hair follicles and lining the digestive system, causing discomfort and illness. Further, depending on the specific type of cancer, chemotherapy is not always effective. This is why recent work in the field of cancer research has focused on picking apart the complex cellular pathways involved in the development of different cancers and identifying the roles of key proteins involved, with the goal of being able to discriminately and effectively eliminate cancerous cells from the body. One of the most prominent of these is the Ras pathway. The term “Ras” collectively refers to 3 proteins (H-Ras, K-Ras, and N-Ras) encoded by a family of proto-oncogenes. Ras proteins are special because they are known to be mutated in as many as 30% of all cancers, especially those of the pancreas and colon.
Despite the prevalence of Ras mutations in a variety of cancers, Ras proteins have stumped the pharmaceutical industry for years. They have proven very difficult to target therapeutically due to their unusually “smooth” shape and lack of “druggable” domains. In other words, Ras proteins naturally lack areas that are accessible, high-affinity binding targets for potential therapeutic agents. Nevertheless, a glimmer of light remains for this elusive target. Recent improvements in computer modeling have enabled researchers to begin discovering and designing molecules that are showing some promise in their ability to inhibit Ras. Stephen Fesik’s group at Vanderbilt University in Nashville, Tennessee has already demonstrated that a number of novel small molecules can bind directly, albeit weakly, to K-Ras and inhibit its activation. The next step may be to follow up on these promising results and customize the small molecules for a higher affinity fit. Kevan Shokat’s group at the University of California, San Francisco, is approaching the Ras problem by exploiting the different mutant versions of Ras that are most prevalent in human cancers, and synthesizing compounds to bind specifically to their mutated residues.
For Ras researchers, one of the most exciting and relatively new resources is the RAS Pathway Clone Collection 2.0. The collection is a set of vectors for use with the Gateway Cloning Platform (Life Technologies, Carlsbad, CA) that represents 180 genes from the Ras signaling pathway. Its major appeal is that it enables the simple construction of vectors to study the function of any of these genes through several different applications such as fluorescent protein fusion studies and the generation of mutant cell lines. Further, for each plasmid in the kit, the transcript most commonly found across all TCGA tumor samples is represented, making the results of any such study as clinically relevant as possible.
Connecting the research community - Addgene's cancer collection pages
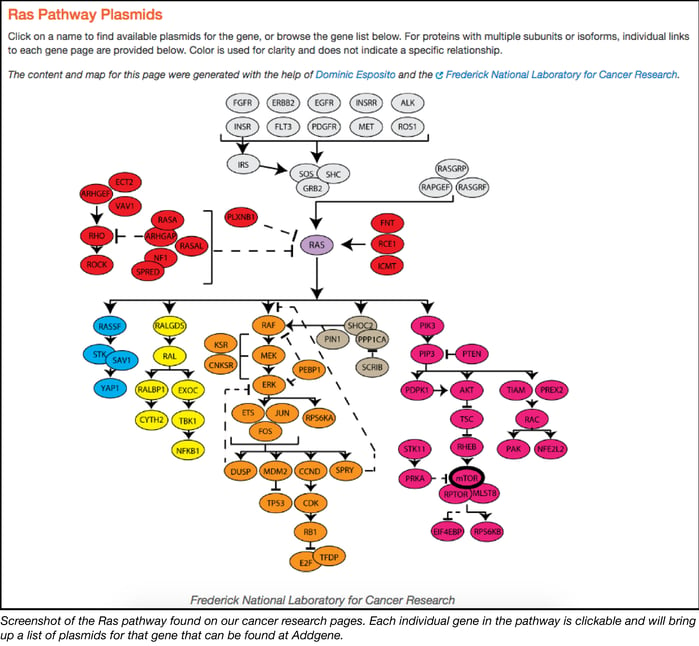
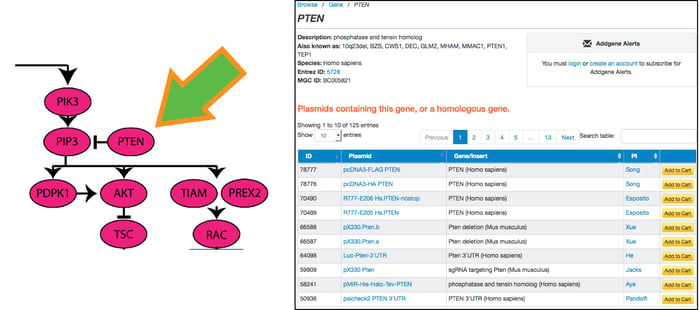
Scientists are making progress. New resources and technologies become available almost daily, yet obstacles remain. One of the biggest challenges facing researchers today is the complexity of the pathways involved in the development of cancer. With hundreds of genes involved in the Ras pathway alone and thousands of investigators studying them, how do we ensure that the research community has access to all of the latest resources? In response to this challenge, Addgene has launched an initiative to curate our collection of cancer-related plasmids such that each is linked to a known major pathway. Now, using our new Cancer Research Plasmids and Resources page, investigators may select a pathway and identify any genes within that pathway that are of interest to their laboratory. A single mouse click on a gene of interest will direct the investigator to a list of all available Addgene plasmids he or she can use to study it (as seen in the screenshot below).
Currently, the page includes interactive maps for the Ras, mTor, and p53 pathways. The curation project continues to evolve rapidly and, with assistance from the research community, will soon expand to include several other important pathways. We invite feedback and contributions from all members of the research community as we continue to to build and develop this new resource! Please leave us your thoughts in the comments section below.
 Michael G. Lemieux is a Scientist at Addgene with interests in education and scientific writing.
Michael G. Lemieux is a Scientist at Addgene with interests in education and scientific writing.
References
1. Martz, Colin A., et al. "Systematic identification of signaling pathways with potential to confer anticancer drug resistance." Science signaling 7.357 (2014): ra121. PubMed PMID: 25538079. PubMed Central PMCID: PMC4353587.
2. Stephen, Andrew G., et al. "Dragging ras back in the ring." Cancer cell 25.3 (2014): 272-281. PubMed PMID: 24651010.
3. Ledford, Heidi. "The ras renaissance." Nature 520.7547 (2015): 278-280. PubMed PMID: 25877186.
4. Prior, Ian A., Paul D. Lewis, and Carla Mattos. "A comprehensive survey of Ras mutations in cancer." Cancer research 72.10 (2012): 2457-2467. PubMed PMID: 22589270. PubMed Central PMCID: PMC3354961.
5. Sun, Qi, et al. "Discovery of small molecules that bind to K‐Ras and inhibit Sos‐mediated activation." Angewandte Chemie 124.25 (2012): 6244-6247. PubMed PMID: 22566140. PubMed Central PMCID: PMC3620661.
Additional Resources
Resources on the Addgene Blog
- Tackling Cancers' Drug Resistance with a New Screening Kit
- Learn How the Clon Tracer Library Can Be Used in Cancer Lineage Tracing
Resources on Addgene.org
Topics: Addgene News, Using Addgene's Website, Cancer





Leave a Comment