This post was contributed by guest blogger Jon Backstrom, a biochemist in the Vanderbilt Eye Institute and Tonia Rex's lab.
A common strategy to determine the binding kinetics of a purified protein involves immobilization on a solid support. This allows washing away of unbound material to calculate the amount of bound ligand (after subtracting out non-specific binding). Historically, glutathione-S-transferase (GST) fusion proteins have been immobilized on a reduced glutathione matrix. The advantage of a fusion protein is the efficient purification of an already immobilized target protein. The disadvantage is that the GST moiety, which forms dimers, may influence binding kinetics of the target ligand. Another important consideration is whether the affinity of an experimental protein-ligand interaction approaches that of GST-glutathione.
Biotin-based immobilization for kinetic analysis
Proteolytic removal of the fusion protein could provide a more relevant kinetic analysis, but the trick is to utilize a second immobilization method. One possibility is covalent modification of the purified target protein with the vitamin biotin. The interaction between biotin and the egg-white protein avidin (or its derivatives streptavidin and neutravidin) is for all practical purposes covalent. Therefore, protein immobilization through biotin-avidin binding could provide a true measurement of high affinity (i.e. sub-nanomolar) protein-ligand interactions. Avidin and its derivatives have four biotin-binding sites per molecule so, theoretically, saturated avidin could form tetramers of biotinylated protein. However, oligomerization of biotinylated proteins on avidin can be controlled easier than formation of GST dimers.
Avi-Tag plasmid for improved biotinylation selectivity
A common strategy to biotinylate proteins involves amine-reactive biotin derivatives that generate covalent bonds with positively-charged amino acids. Unfortunately, the location of biotin incorporation and to a lesser extent, the number of added biotins per molecule cannot be well-controlled. The lack of site selectivity typically diminishes the apparent affinity for the ligand, especially if the modified amines are located near the ligand binding site. A selective approach entails the use of the enzyme biotin ligase, which binds to a small (< 20 amino acid) AviTag sequence in a protein and adds biotin to a lysine residue within the AviTag. However, large amounts of the target protein are typically required due to the relatively low efficiency of biotinylation, at least in vitro.
Our solution was to exploit the best of both worlds. We generated a plasmid that produces biotin ligase and a protein of interest fused to maltose binding protein (MBP, New England BioLabs). MBP has a similar function as GST in that it provides a method to express a foreign protein in bacteria and subsequently purify the fusion protein from a bacterial extract. The MBP tag can be removed using Xa protease. The carboxyl-terminus of the fusion protein includes an AviTag and a 6X HisTag for affinity purification (see plasmid map and cartoon below). Including biotin in your culture and inducing protein expression with IPTG results in in vivo biotinylation. The co-expressed ligase will biotinylate the lysine residue in the AviTag. As a side note, we found that adding a TEV linker dramatically increased accessibility of the AviTag and HisTag, which were “buried” in a few of our proteins that lacked TEV. Although we were unable to cleave TEV within the protein we tested, it may be possible with other fusion proteins - but it would remove both the AviTag and HisTag.
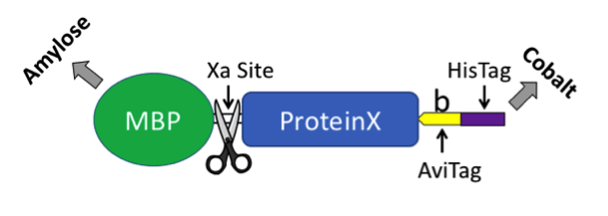
We were surprised that we couldn’t find an all-in-one plasmid that provided an easy way to biotinylate a fusion protein in bacteria. Our initial attempt to create a plasmid based on the biotin ligase BirA backbone (Eric Campeau, Addgene plasmid #26624) was unsuccessful. Nothing more joyful that getting to the final step and the transformed bacteria refuse to grow. Eventual success was achieved by inserting BirA into a pMal plasmid (NEB) thus creating the pMal-T-Avi-His/BirA plasmid now available through Addgene.
We have successfully used this plasmid to biotinylate the extracellular domain of the erythropoietin receptor and subsequently evaluated interactions with mutant and wild-type erythropoietin molecules. The in vivo biotinylation and purification strategy outlined here provides a basis to determine binding kinetics using a multitude of methods including surface plasmon resonance (Biacore, GE Life Sciences) or bio-layer interferometry (Octet, Pall ForteBio).
Tips for using pMal-T-Avi-His/BirA to add biotin to your protein of interest
- Grow fusion protein plasmid in NEB T7 SHuffle bacteria (C3029J). We are clearly NEB “fanboys” as the bacteria significantly enhance protein folding (tested using real science).
- Make a stock solution of 2.5 mM biotin (50X) because 5 mM is barely soluble at RT.
- Track expression and purification of biotinylated protein on blots with Streptavidin-phosphatase (Jackson ImmunoResearch, 016-050-084; 0.2 μg/ml final). Both rabbit (EMD, AB3596) and mouse (NEB, E8032S) antibodies against MBP are commercially available.
- Factor Xa loves calcium, but metal-chelate matrices, not so much. Maybe a calcium-independent proteinase site could be used for plasmid version 2.0?
- Instruments such as an Octet may require less in vivo-biotinylated protein (total amount) relative to proteins that have been biotinylated using less efficient methods.
Many thanks to our guest blogger Jon Backstrom.
 Jon Backstrom, Ph.D is a biochemist in the Vanderbilt Eye Institute and is particularly interested in cell signaling.
Jon Backstrom, Ph.D is a biochemist in the Vanderbilt Eye Institute and is particularly interested in cell signaling.
Additional Resources on the Addgene Blog
- Check out other Hot Plasmids
- Find Additional Posts on Mol Bio Techniques
- Find Additional Posts on Plasmid Technologies
Resources at Addgene.org
- Learn about Popular Plasmids in the Repository
- Learn about New Plasmid Technologies in our Hot Plasmid Articles
Topics: Other Plasmid Tools, Plasmids






Leave a Comment