Update (November 18, 2016): Researchers from a variety of institutions recently reported their inability to recapitulate the results of Gao et al 2016 in a letter to Protein & Cell.
Update (August 3rd, 2017) THE ORIGINAL NgAgo ARTICLE DISCUSSED IN THIS POST HAS BEEN RETRACTED AND FOLLOW UP STUDIES HAVE FAILED TO DEMONSTRATE GENOME EDITING BY THIS TOOL
This post was contributed by guest blogger Pooran Dewari. Any views in this post are those of the guest blogger and do not necessarily represent the views of Addgene. Addgene performs Sanger sequencing on select regions of all distributed plasmids as part of quality control, but does not perform functional tests.
The newest genome engineer sharing the stage with much-lauded CRISPR-Cas9 is DNA-guided endonuclease NgAgo! We'll discuss how NgAgo is faring with users in a minute, but, to start, let's review why NgAgo is in the spotlight and take a moment to remember that NgAgo has only been available for genome editing for a few months. More time is required for its optimization and development before it can truly be pitted against CRISPR head-to-head.
Exciting features of NgAgo
First, unlike Cas9, NgAgo does not require a PAM sequence for target recognition, which gives a researcher unprecedented freedom to target any sequence of DNA. Second, NgAgo target specificity is dictated by a DNA guide sequence rather than an RNA guide as with Cas9. Third, it can be used to target “difficult” GC-rich regions of the genome, which seem resistant to Cas9 cleavage (1).
Both the CRISPR-Cas9 and NgAgo genome engineering methods have their own pros and cons (2). The NgAgo DNA guide is inexpensive and can be 5’-phosphorylated in house using a simple PNK reaction or can be purchased 5’-modified from a company. However, unlike an RNA guide, it cannot be produced from a plasmid in cells. Additionally, in vitro assembly of NgAgo/ssDNA requires incubation at 55 °C - a dangerous, non-physiological temperature for mammalian cells and one which diminishes the endonuclease activity of the enzyme. In vivo, only nascent NgAgo protein (probably while it is being synthesized) can form a complex with an ssDNA guide. Therefore, researchers must co-transfect cells with 5’-P-ssDNA guides and an NgAgo expression plasmid to edit a gene in vivo.
On the other hand, the widespread success of CRISPR-Cas9 across multiple model organisms (3) lies in the versatility of its delivery methods. Researchers can conveniently transfect/inject cells with in vitro preassembled Cas9/gRNA complexes or with guide RNA- and Cas9-expression plasmids. Cas9 guide RNA delivery does however rely on cloning into a vector or in vitro synthesis, which could be less desirable if one has to target multiple sites in a single experiment. Also, the NGG PAM sequence might not always be available at an optimal distance from the target site. For example, NGG may not be present in AT-rich regions of the genome or just after a stop codon (as required for C-terminus epitope tagging).
NgAgo survey
Since it was first published in Nat Biotech in May 2016 (1), NgAgo has received much attention and the plasmid has been requested by labs around the globe nearly 400 times. Researchers have been excitedly testing NgAgo's genome editing applications. So, how does NgAgo stack up thus far? A few weeks ago, I was going through NgAgo google forums where people write their experiences with the new method and look for advice/tips. Based on the discussions in the forums, it appeared that many researchers were having difficulty achieving indel formation (gene disruption) with this new technology. To dig more into what researchers felt about NgAgo, I conducted a survey and asked researchers if they had tested and compared NgAgo head-to-head with CRISPR-Cas9. The survey had two sections, the first section asked which of the two methods researchers found to be more successful, and the second section asked basic questions about the NgAgo protocols used by respondents (transfection method, amount of each component per transfection etc.).
Survey results
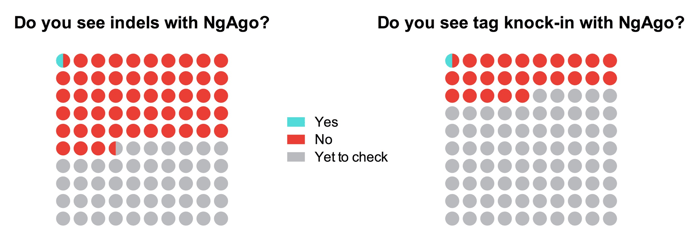
As of August 1, 2016, a total of 165 researchers responded to the survey. When asked if they could detect indels (an indication of gene disruption ability) with NgAgo, one respondent out of 88 confirmed successful indel formation at a target site. Also, 41 respondents attempted to use the technique for epitope tagging but only one of them could detect knock-in of tags at a target site. Overall, 64% of researchers were unhappy with the new method and 65% wanted to wait until someone else optimizes an NgAgo protocol for their favorite model system. The vast majority of the respondents (70%) used a lipofectamine-based method to deliver NgAgo ingredients into the cell, the same method used by Gao et al, 2016 (1).
View the Complete and Up-to-Date Results of Pooran's NgAgo Survey
The take-home message
While most of the respondents have been struggling to achieve genome editing with NgAgo, this does not mean the technology is dead in the water. On the bright side, successful indel generation and epitope knockin was achieved by a few respondents suggesting that NgAgo can indeed be optimized for genome editing in mammalian cells. However, the fact that most of the researchers surveyed here could not achieve indels with the NgAgo system indicates that it is very tricky to use and fussy to optimize. One reason could be the instability of the supposedly alien 5’-phosphorylated ssDNA guides, which might be rapidly degraded by the cellular machinery, leaving a small amount of active NgAgo/ssDNA complex in the cell. As proposed in the original article, to achieve greater efficacy, researchers will have to carry out repeated transfections with the ssDNA guide, although many may find this inconvenient. Another possible reason for the trickiness of NgAgo is the inability of mature NgAgo protein to load an ssDNA guide. It is possible that, during translation, only a small fraction of nascent NgAgo protein molecules manages to form a functional complex with ssDNA leading to inefficient downstream editing.
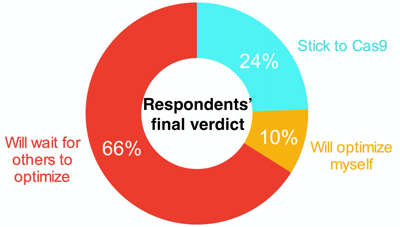
Overall, the real take-home message thus far, based on survey respondents "Final Verdict", is that more time is required for NgAgo optimization before we can make any major conclusions:
That said, many people are having difficulty with the NgAgo system. I encourage you to share your results and protocols with the research community if you've had success with it. Some great places to get your results out there include:
- Google Groups - I've specifically set up a forum where NgAgo users can share protocols and tips
- Twitter - Tweet suggestions and tips with #NgAgo
- The Addgene Blog - Leave suggestions and tips in the comments section below or contact blog@addgene.org if you'd like to write a more detailed post
It may take a while to optimize NgAgo for your favorite model system, but the genome-editing field is advancing rapidly and it wouldn’t be a surprise if someone came up with a mutant NgAgo (or a novel endonuclease) protein that can form a complex with a 5’-P-ssDNA guide at room temperature. Until that time, those who have been lucky with NgAgo should explicitly share tips and protocol details with the scientific community on NgAgo forums, blogs, or in the comments section below. Keep pipetting and happy genome editing!
Check Out Other Argonaute Plasmids from the Doudna Lab
Many thanks to our Guest Blogger Pooran Dewari!

Pooran Dewari is a postdoc at the MRC Centre for Regenerative Medicine (CRM), Edinburgh. His research interests include optimizing genome-editing methods in brain cells and understanding brain tumor biology. When not snipping DNA, he tries random things (101 lessons on Spanish, C++, guitar, photography) and often dreams of a sunny day in Edinburgh.
References
1. Gao, Feng, et al. "DNA-guided genome editing using the Natronobacterium gregoryi Argonaute." Nature biotechnology (2016). PubMed PMID: 27136078.
2. Burgess, Shawn., et al. "Questions about NgAgo." Proteins & Cell (2016).
3. "NgAgo: Genome-editing by a DNA-guided enzyme." DNA-scissors (blog). Blog website.
4. Hsu, Patrick D., Eric S. Lander, and Feng Zhang. "Development and applications of CRISPR-Cas9 for genome engineering." Cell 157.6 (2014): 1262-1278. PubMed PMID: 24906146. PubMed Central PMCID: PMC4343198.
Resources on the Addgene Blog
- Learn More about NgAgo
- Enhance CRISPR Targeting Specificity with eSpCas9 and SpCas9-HF1
- See How Cpf1 Stacks Up to Cas9 and NgAgo
Resources on Addgene.org
- Request NgAgo
- Find CRISPR Resources
- Catch Up on Your CRISPR Background
Topics: CRISPR, Cas Proteins





Leave a Comment