Last updated Oct 7, 2020 by Gabrielle Clouse.
This post was contributed by guest bloggers Marcelle Tuttle and Alex Chavez, researchers at the Wyss Institute for Biologically Inspired Engineering.
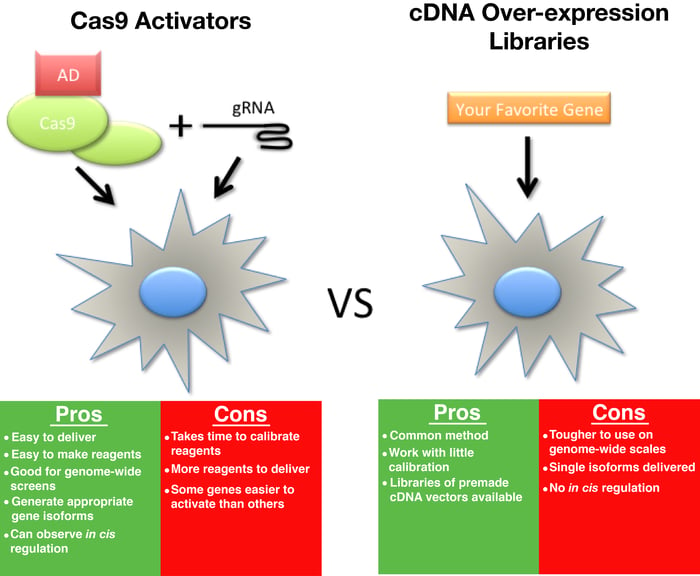
CRISPR/Cas9 is an enormously plastic tool and has taken the scientific world by storm. While Cas9 has been most widely used to create specific edits in DNA, there has also been significant work on constructing Cas9 transcriptional activators. These constructs allow for the upregulation of essentially any gene by fusing mutants of Cas9 deficient in DNA cutting activity to a transcriptional activation domain (Fig 1).
Two ways to use CRISPRa for your research
CRISPRa for specific gene targets
CRISPR activation for genes with known functions has the potential to be used in therapeutics, as it can increase gene expression at certain loci to create changes in phenotypes. For example, CRISPR activation has been used to differentiate induced pluripotent stem cells (Chavez et al., 2015) into neuronal cells by activating genes that cause differentiation and to reverse HIV viral latency (Bialek et al., 2016).
Additionally, CRISPR-based activation can be made inducible upon environmental triggers. This is done by creating a split dCas9 protein that can reassemble when exposed to specific UV wavelengths or chemicals (Polstein and Gersbach, 2015; Zetsche et al., 2015). These types of CRISPR activation are also reversible: once the environmental trigger leaves, gene expression is no longer activated.
However, there are several factors that may prevent your gene from being up-regulated. For instance, generally, the more highly expressed a gene is under native conditions, the less activation you can achieve using a CRISPRa. Your gene of interest might already be hitting an upper bound of activation that current Cas9 systems cannot help you pass. In addition, activation experiments often require quite a bit of tuning before you know your system is working as expected. Finally, for each gene you want to activate, you should also be ready to test three or four guides directed towards that gene as there can be a large difference in guide potency.
Thus, in cases where users have a single gene they want to activate, we would recommend using a cDNA overexpression vector rather than going through all the troubleshooting required for Cas9-based activation.
CRISPRa for genome-wide studies
One of the best uses for Cas9 activators is in genetic screening. gRNAs targeting every gene in the human genome for example, can be made easily and cheaply using oligo library synthesis. Prior to Cas9 activators, similar tools were made using other DNA binding proteins such as zinc fingers (ZF) and TAL effectors (TALE). Unlike these constructs, however, Cas9 allows you to easily change the sequence targeted by the activator by simply providing a new gRNA rather than engineering an entirely new protein. This makes it much cheaper to use Cas9 activators.
This larger scale of activation allows for exploratory-based research as well. For example, CRISPRa can be used to find potential cancer drug targets by finding genes that cause cell death in cancer cell lines upon activation (Gilbert et al., 2014; Behan et al., 2019). Because of its ability to target multiple gene loci simultaneously, CRISPRa can also be used to map molecular pathways and genetic interactions. Additionally, large scale CRISPR-based screens can be used to gain insight into protein structure and function (Pan et al., 2018).
cDNA libraries, which consist of plasmids that over-express coding sequences from a given cell type or organism, have been used in a similar manner to Cas9 activators. However, these can be difficult to construct and deliver when compared to gRNAs. Additionaly, cDNAs cannot be used to study in cis regulation and also suffer from an inability to easily deliver the appropriate isoform(s) of a given gene, as, many times, the isoform(s) common to a particular cell type are unknown or not readily available. By activating from the native context of the gene, Cas9 activators efficiently solve these problems.
Which Cas9 activator should I use?
There are a wide variety of activators you can use for your experiments. We have found that SAM (Konermann et al., 2014), Suntag (Tanenbaum et al., 2014, Gilber et al, 2014), and VPR (Chavez et al., 2015)) are good choices across multiple cell lines (HEK293T, MCF7, U2-OS, Hela, N2A, 3T3) and organisms (Chavez et al., 2016). To learn more about the different types of Cas9 activators, check out our blog post covering VP64, Suntag, SAM, and VPR.
Worry less about off targets
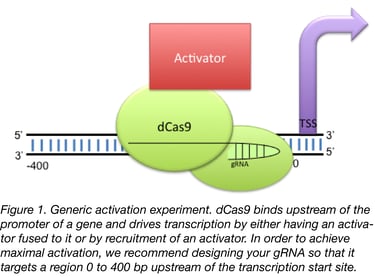
Unlike Cas9 cutting activity, off target effects are generally not regarded as being a large problem for Cas9 activators. This is believed to be true given the results of previous RNA-seq experiments (Konermann et al., 2014, Chavez et al., 2014, Chavez et al., 2016) along with a belief that the odds are very low that Cas9 would have an off-target that lands in the promoter of another gene, thereby driving aberrant transcription. That being said, we generally pick guides by putting the promoter of the gene into a gRNA finder such as WU-CRISPR (Wong et al., 2015)) or our lab’s sgRNA scorer 1.0 (Now 2.0) (Chari et al., 2015) and picking whichever guides are closest to the transcription start site (TSS). We recommend targeting the guides to a region less than 200 bp upstream of TSS for best results but up to 400 bp works reasonably well.
Best of luck on your experiments!
Many thanks to our guest bloggers Marcelle Tuttle and Alex Chavez!

Marcelle Tuttle is a medical student at Tufts University and former researcher at the Wyss Institute for Biologically Inspired Engineering.
 Alejandro (Alex) Chavez is a member of the Addgene Scientific Advisory Board. His laboratory at Columbia University is focused advancing scientific discovery through the creation of innovative technologies. To date his group has develop several popular CRISPR-based tools for altering DNA sequence or regulating gene expression.
Alejandro (Alex) Chavez is a member of the Addgene Scientific Advisory Board. His laboratory at Columbia University is focused advancing scientific discovery through the creation of innovative technologies. To date his group has develop several popular CRISPR-based tools for altering DNA sequence or regulating gene expression.
References
Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, Santos R, Rao Y, Sassi F, Pinnelli M, Ansari R, Harper S, Jackson DA, McRae R, Pooley R, Wilkinson P, van der Meer D, Dow D, Buser-Doepner C, Bertotti A, Trusolino L, Stronach EA, Saez-Rodriguez J, Yusa K, Garnett MJ (2019) Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 568:511–516 . https://doi.org/10.1038/s41586-019-1103-9
Bialek JK, Dunay GA, Voges M, Schäfer C, Spohn M, Stucka R, Hauber J, Lange UC (2016) Targeted HIV-1 Latency Reversal Using CRISPR/Cas9-Derived Transcriptional Activator Systems. PLoS ONE 11:e0158294 . https://doi.org/10.1371/journal.pone.0158294
Chari R, Mali P, Moosburner M, Church GM (2015) Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods 12:823–826 . https://doi.org/10.1038/nmeth.3473
Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, Braff JL, Davidsohn N, Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, Church GM (2015) Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12:326–328 . https://doi.org/10.1038/nmeth.3312
Chavez A, Tuttle M, Pruitt BW, Ewen-Campen B, Chari R, Ter-Ovanesyan D, Haque SJ, Cecchi RJ, Kowal EJK, Buchthal J, Housden BE, Perrimon N, Collins JJ, Church G (2016) Comparison of Cas9 activators in multiple species. Nat Methods 13:563–567 . https://doi.org/10.1038/nmeth.3871
Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS (2014) Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159:647–661 . https://doi.org/10.1016/j.cell.2014.09.029
Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F (2014) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517:583–588 . https://doi.org/10.1038/nature14136
Pan J, Meyers RM, Michel BC, Mashtalir N, Sizemore AE, Wells JN, Cassel SH, Vazquez F, Weir BA, Hahn WC, Marsh JA, Tsherniak A, Kadoch C (2018) Interrogation of Mammalian Protein Complex Structure, Function, and Membership Using Genome-Scale Fitness Screens. Cell Systems 6:555-568.e7 . https://doi.org/10.1016/j.cels.2018.04.011
Polstein LR, Gersbach CA (2015) A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 11:198–200 . https://doi.org/10.1038/nchembio.1753
Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD (2014) A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell 159:635–646 . https://doi.org/10.1016/j.cell.2014.09.039
Wong N, Liu W, Wang X (2015) WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol 16: . https://doi.org/10.1186/s13059-015-0784-0
Zetsche B, Volz SE, Zhang F (2015) A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol 33:139–142 . https://doi.org/10.1038/nbt.3149
Additional Resources on the Addgene Blog
- Learn how to Choose the Best Cas9 Variant for Your Next Experiment
- Find out More about Genome-wide Screens with CRISPR/Cas9
- Learn How CRISPR Has Been Used with AAV
Resources on Addgene.org
- Find Cas9 Activators
- Find Cas9 Repressors
- Catch Up on Your CRISPR Background with Our CRISPR Guide Pages
Topics: CRISPR, Cas Proteins






Leave a Comment