This post was contributed by guest bloggers Erik L. Snapp and Lindsey M. Costantini.
"You underestimate the power of the Dark Side."
--Darth Vader in "Return of the Jedi"
While Vader was referring to the evil side of a mystical "Force," this quote is equally applicable to many microscopy experiments with fluorescent proteins (FPs) localized to compartments other than the cytoplasm. That is, unfortunately, some investigators realize too late that they have missed the impact of dark, non-fluorescent, and misfolded FP-fusions on quantitative imaging experiments and cell physiology in general.
 Pitfalls of Fluorescent Protein Fusions
Pitfalls of Fluorescent Protein Fusions
Observing a bright signal for the first time after cloning an FP fusion with your favorite protein of interest is exciting and rewarding. But, what if an FP fusion with a resident endoplasmic reticulum (ER) protein instead localizes to the cytoplasm? Or more troubling, what if a significant fraction of the fusion protein molecules fail to correctly fold? Misfolded FPs do not fluoresce. A dark population is not readily apparent and can confound quantitative imaging experiments or even negatively impact cells (see Figure 1).
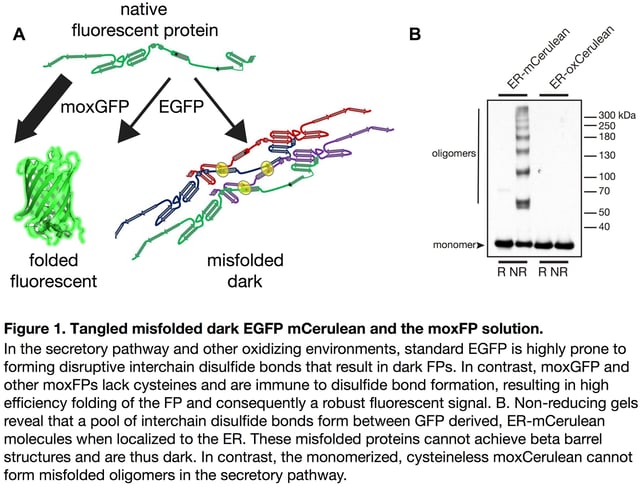
When behaving properly, FPs enable researchers to investigate the localization and dynamics of fluorescent protein fusions in live cells in real time. Previously, we have described a number of practical considerations that should be taken into account when deciding where to place an FP within a fusion construct [1,2], i.e. positioning of the FP sequence relative to cell compartment targeting sequences, which often have absolute position requirements. For example, a KDEL ER retrieval sequence only functions at the extreme C-terminus [3]. You must therefore determine your cloning strategy and decide whether you will create an N- or C-terminal fusion based on your experimental needs.
Equally important, before cloning you must consider whether and how well your FP will function in the environment where it will localize [4]. Most investigators are surprised to learn that FPs are often poorly suited for cellular compartments other than the cytoplasm. Generally, FPs evolved in or were engineered for use in cytoplasmic environments. However, approximately 40% of human (and most eukaryotic) proteins localize to chemically distinct subcellular environments, including the organelles that compose the secretory pathway, endocytic vesicles, mitochondria, lysosomes or they are secreted into the extracellular milieu. Many of the resident proteins within these compartments undergo significant post-translational modifications including glycosylation, disulfide bond formation, and proteolytic cleavage. FPs localized to organelles are equally susceptible to these non-native modifications, which can all potentially affect function [5]. We and others have reported that FP cysteine residues form inappropriate disulfide bonds in the secretory pathway (Figure 1) [4,6–8]. Additionally, FPs that encode N-glycosylation consensus sequences are modified with the addition of N-glycans [9]. Both of these posttranslational modifications can disrupt FP folding rendering the protein non-fluorescent.
Unfortunately, accumulated misfolded and nonfunctional (dark) fluorescent proteins cannot be ignored. They have the potential to interfere with the function of resident ER chaperones and may impact cellular function or possibly viability. FP misfolding can even disrupt fusion protein localization. For example, the formation of inappropriate inter-chain disulfide bonds between FPs blocks Golgi complex-localized fusions from properly exiting the ER and fluorescing (see Figure 2) [5]. When using FPs containing two cysteine residues and a sequence intended to localize them to the secretory pathway, a dark pool that is mislocalized to the ER can be detected with immunofluorescence. This population of dark fusions is likely misfolded, blocked from proceeding through the secretory pathway, and retained in the ER.
FPs have been undeniably powerful and successful tools that have enabled numerous cell biology assays. However, there has been a pervasive view that most FPs in a cell will fold and that the fluorescence pattern is equivalent to the distribution of FP-fusion proteins. Our studies and those of others (5, 6, 7, 10, 11) highlight the very real consequences of misfolded dark FP fusions including erroneous quantitation of fusion protein levels. It remains unclear whether the FP misfolding impacts fusion protein function and this could lead to under and overestimation of fusion protein activity. We encourage investigators to characterize FP-fusion proteins functionally relative to the untagged protein of interest. To avoid the accumulation of dark fluorescent proteins in your experiments, please consider the following:
How can one ensure that an FP fusion with a non-cytosolic protein functions properly?
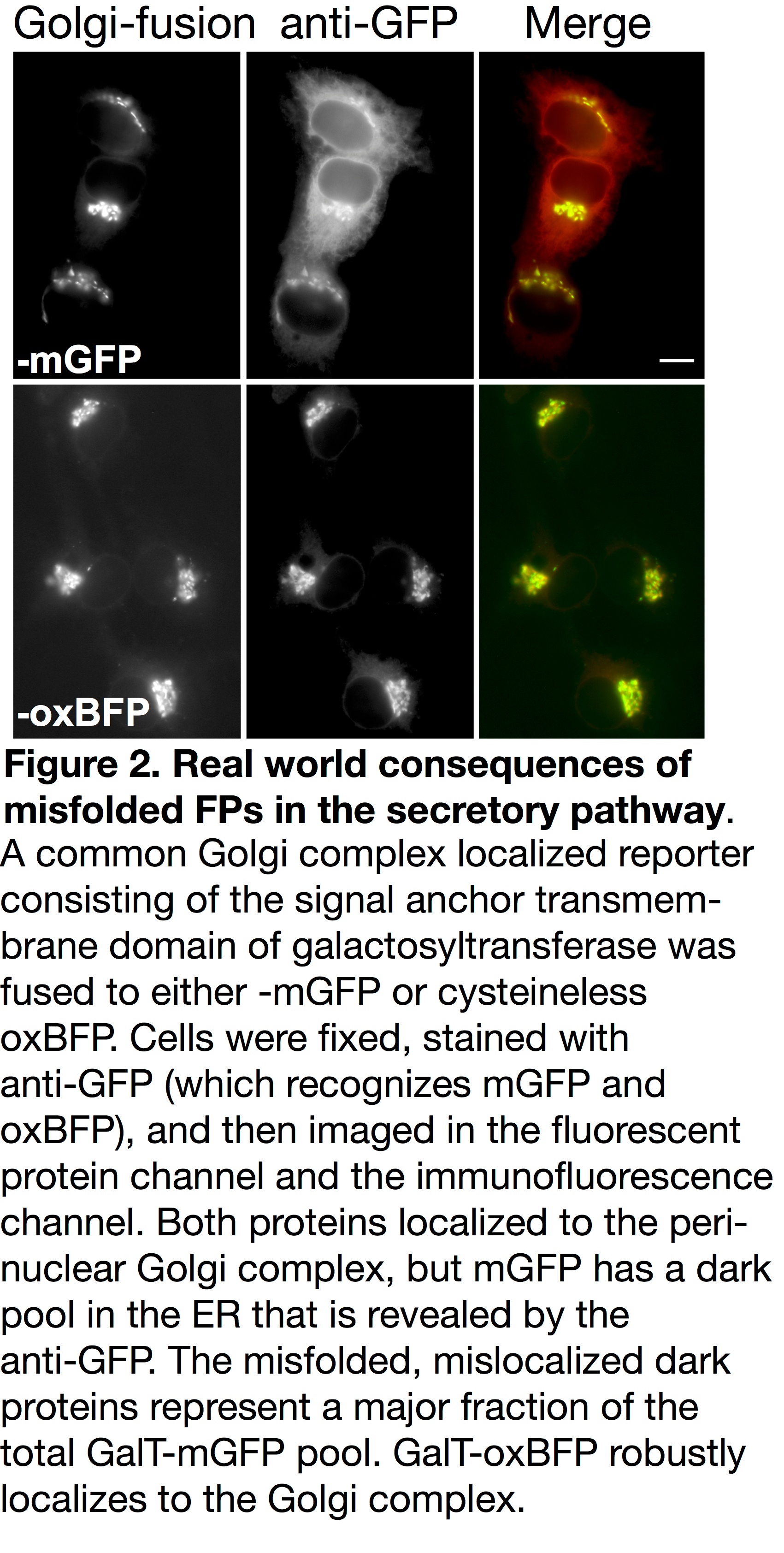 We suggest starting by using the newly published oxidative optimized palette of FPs (moxFPs) [5]. The moxFPs overcome the inherent problems of popular fluorescent proteins and were re-engineered by mutating cysteine residues and N-glycosylation consensus sequences in popular FPs. This generated a palette of inert green, cyan, yellow, and blue variants that can be used in various combinations. For example, with standard fluorescent microscope filter sets, users can label multiple proteins of interest with a combination of green and blue or cyan and yellow moxFPs. The inherent brightness and spectral characteristics of the moxFPs are comparable to the non-optimized parental proteins. Our recent publication illustrated the quantitative increase in fluorescent signal achieved when utilizing an optimized moxFP as opposed to standard FPs. The increase in signal is not due to a brighter FP, but rather because of the correct folding and fluorophore formation by most, if not all, of the FP fusions.
We suggest starting by using the newly published oxidative optimized palette of FPs (moxFPs) [5]. The moxFPs overcome the inherent problems of popular fluorescent proteins and were re-engineered by mutating cysteine residues and N-glycosylation consensus sequences in popular FPs. This generated a palette of inert green, cyan, yellow, and blue variants that can be used in various combinations. For example, with standard fluorescent microscope filter sets, users can label multiple proteins of interest with a combination of green and blue or cyan and yellow moxFPs. The inherent brightness and spectral characteristics of the moxFPs are comparable to the non-optimized parental proteins. Our recent publication illustrated the quantitative increase in fluorescent signal achieved when utilizing an optimized moxFP as opposed to standard FPs. The increase in signal is not due to a brighter FP, but rather because of the correct folding and fluorophore formation by most, if not all, of the FP fusions.
Our moxFPs currently represent one of the most inert solutions for imaging in a variety of cellular compartments including the secretory pathway (Figure 2), the inner membrane space of mitochondria and chloroplasts, the extracellular milieu, and gram negative bacteria periplasm. Efforts to tag endogenous genes for proteins in these compartments using CRISPR systems should strongly consider using moxFPs over standard FPs for the reasons listed above. In addition, because the moxFPs are highly monomeric, they are an excellent choice for fusions with integral membrane proteins and membrane associated proteins (i.e. GPI-anchored proteins), which are more susceptible to the oligomerizing effects of many standard FPs. The moxFPs are now available through Addgene
Many thanks to our guest bloggers Erik L. Snapp and Lindsey M. Costantini!
 Erik Lee Snapp received his PhD in Molecular Microbiology and Immunology from Oregon Health Sciences University, did his postdoctoral fellowship with Jennifer Lippincott-Schwartz at the National Institutes of Health and he is currently an Associate Professor at Albert Einstein College of Medicine in the Department of Anatomy and Structural Biology. His interests include the quality control of secretory proteins in the Endoplasmic Reticulum and live cell imaging approaches. Erik is also an avid long distance runner, gardner, and cook.
Erik Lee Snapp received his PhD in Molecular Microbiology and Immunology from Oregon Health Sciences University, did his postdoctoral fellowship with Jennifer Lippincott-Schwartz at the National Institutes of Health and he is currently an Associate Professor at Albert Einstein College of Medicine in the Department of Anatomy and Structural Biology. His interests include the quality control of secretory proteins in the Endoplasmic Reticulum and live cell imaging approaches. Erik is also an avid long distance runner, gardner, and cook.
 Lindsey M. Costantini received her PhD in Biomedical Research from Albert Einstein College of Medicine. Her doctoral research was conducted in the laboratory of Erik Lee Snapp and focused on optimizing fluorescent proteins for subcellular environment. Currently, she is a SPIRE postdoctoral fellow at the University of North Carolina at Chapel Hill. Under the co-mentorship Blossom Damania and Jack Griffith, her research focuses on the replication machinery of Kaposi’s sarcoma-associated herpesvirus.
Lindsey M. Costantini received her PhD in Biomedical Research from Albert Einstein College of Medicine. Her doctoral research was conducted in the laboratory of Erik Lee Snapp and focused on optimizing fluorescent proteins for subcellular environment. Currently, she is a SPIRE postdoctoral fellow at the University of North Carolina at Chapel Hill. Under the co-mentorship Blossom Damania and Jack Griffith, her research focuses on the replication machinery of Kaposi’s sarcoma-associated herpesvirus.
References
1. Snapp, Erik. "Design and use of fluorescent fusion proteins in cell biology."Current Protocols in Cell Biology (2005): 21-4. Pubmed PMID: 18228466. Pubmed Central PMCID: PMC2875081.
2. Snapp, Erik Lee. "Fluorescent proteins: a cell biologist's user guide." Trends in cell biology 19.11 (2009): 649-655. Pubmed PMID: 19819147. Pubmed Central PMCID: PMC2784028.
3. Munro, Sean, and H. R. Pelham. "Use of peptide tagging to detect proteins expressed from cloned genes: deletion mapping functional domains of Drosophila hsp 70." The EMBO journal 3.13 (1984): 3087. Pubmed PMID: 6526011. Pubmed Central PMCID: PMC557822.
4. Costantini, Lindsey M., and Erik Lee Snapp. "Fluorescent proteins in cellular organelles: serious pitfalls and some solutions." DNA and cell biology 32.11 (2013): 622-627. Pubmed PMID: 23971632. Pubmed Central PMCID: PMC3806368.
5. Costantini, Lindsey M., et al. "A palette of fluorescent proteins optimized for diverse cellular environments." Nature communications 6 (2015). Pubmed PMID: 26158227. Pubmed Central PMCID: PMC4499870.
6. JAIN, R., et al. "Oligomerization of green fluorescent protein in the secretory pathway of endocrine cells." Biochem. J 360 (2001): 645-649. Pubmed PMID: 11736655. Pubmed Central PMCID: PMC1222268.
7. Aronson, Deborah E., Lindsey M. Costantini, and Erik L. Snapp. "Superfolder GFP is fluorescent in oxidizing environments when targeted via the Sec translocon." Traffic 12.5 (2011): 543-548. Pubmed PMID: 21255213. Pubmed Central PMCID: PMC3079558.
8. Suzuki, Takahisa, et al. "Development of cysteine-free fluorescent proteins for the oxidative environment." PloS one 7.5 (2012): e37551. Pubmed PMID: 22649538. Pubmed Central PMCID: PMC3359384.
9. Costantini, Lindsey M., et al. "Cysteineless non-glycosylated monomeric blue fluorescent protein, secBFP2, for studies in the eukaryotic secretory pathway."Biochemical and biophysical research communications 430.3 (2013): 1114-1119. Pubmed PMID: 23257162. Pubmed Central PMCID: PMC3552020.
10. Paladino, Simona, et al. "Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins." The Journal of cell biology 167.4 (2004): 699-709. Pubmed PMID:15557121. Pubmed Central PMCID: PMC2172584.
11. Jokitalo, Eija, et al. "Golgi clusters and vesicles mediate mitotic inheritance independently of the endoplasmic reticulum." The Journal of cell biology 154.2 (2001): 317-330. Pubmed PMID: 11470821. Pubmed Central PMCID: PMC2150754.
Resources Avaliable at Addgene- Read Our Blog Post on Protein Tags
- Read Other Posts on Fluorescent Proteins
- Browse Our Fluorescent Protein Pages
- Find the mOX Fluorescent Proteins at Addgene
Topics: Fluorescent Proteins, Generating Fusions






Leave a Comment