Updated May 10, 2021.
In the world of fluorescent proteins and their use for imaging cell biology, Michael Davidson’s lab at Florida State University has been the go-to place. In 2012, his National High Magnetic Field Lab worked with an impressive 1,350 scientists from more than 275 institutions all over the world. In the course of all those collaborations over the years, he and his colleagues built a Molecular Expressions collection including some 3,300 plasmids along with image galleries and educational resources to go with them. This collection of plasmids is available in an easily searchable format on Addgene’s Michael Davidson Fluorescent Protein Collection webpage.
These plasmids are divided into the following categories:
- Over 300 empty backbones with 100+ distinct fluorophores to choose from in the green, red, yellow, cyan, blue, and orange spectra.
- A comprehensive list of cell markers all tagged with mEmerald.
- Plasmids to monitor protein-protein interactions through multicolor labeling and FRET.
- Specialty constructs for biosensing.
Over 300 plasmid backbones spanning a rainbow of fluorescent proteins
“It’s a great collection with over 300 backbones alone,” said Addgene’s Lianna Swanson, who has been working with members of Davidson's lab to coordinate the impressively big deposit. “He has every fluorescent protein under the sun, from the standard oldies but goodies (e.g., EGFP and YFP) to the new and improved fruit colors (e.g., apple, papaya, and tomato) and the photoactivatable fluors (e.g. Phamret and Dendra). It’s just such a great collection with such variety.”
In most cases, backbones are available to attach the gene of interest to either the N-or the C-terminus. Even if a researcher’s gene of interest isn’t in the ORF collection, the fluorescent proteins are, which makes mixing and matching a relatively easy proposition, explained Michelle Baird, who has created plasmids as a technician in the Davidson lab for the last four years.
“Researchers can make their own target if it is not already made for them,” Baird said. “They are all designed in the same vector backbone with very similar restriction enzymes. You can essentially cut and paste quite quickly. Researchers should keep in mind that a very simple sequential or double restriction enzyme digest will open up possibilities exponentially as far as new targets and fluorescent proteins are concerned.”
For more information on choosing a backbone and making fusions, see this blog post from Joachim Goedhart: Fluorescent Proteins 101: GFP Fusion Proteins - Making the Right Connection.
mEmerald plasmids and many more!
Baird has a helpful tip for anyone looking to peruse the collection without getting overwhelmed by its scale: Look at the mEmerald variants first, followed by the color variants. The mEmerald plasmids offer a comprehensive list of readily available targets since the Davidson lab always tagged new proteins with mEmerald first, Baird says.
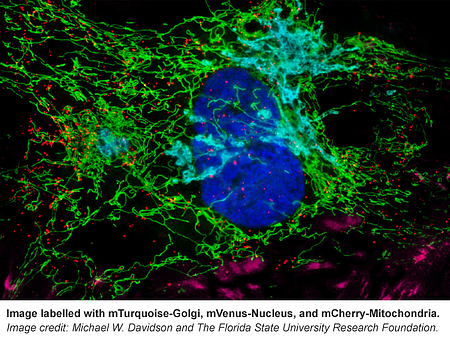
From F-Actin to mitochondria, from the nucleus to cell membrane, this collection has fluorescently tagged versions of many proteins known to localize to specific cellular structures.The Davidson lab even included some of their own reference images that can be found on the corresponding plasmid pages (e.g. see Supplemental Documents here). If you’re not sure where your protein of interest localizes in the cell, use these markers as your guide.
As Michael Davidson told the Tallahassee Democrat in a story about his life’s work and the stage IV lung cancer that has now prompted him to deposit the tools he and his team created with the research community at large, "[Fluorescent proteins are] a whole new way of seeing cell biology, seeing what interacts with what, what affects what. It's like fixing a Maserati. If we know how all the parts go together, we can fix it. Right now, we have no idea."
Now, it will be that much easier for scientists everywhere to get a clearer picture of that intracellular world, even as the Davidson lab prepares to shutter its doors.
“We wanted to give all of these vectors to the scientific community and contribute in a way that everyone could utilize what we had made,” Baird said. “It won’t sit in a freezer at FSU. Mike is really the one who had been very adamant that we distribute to as many researchers as we could, and Addgene is a wonderful vehicle for us to do that.” At Addgene, we are equally excited to be helping the Davidson lab to share their reagents. "We are proud to be able to bring such an important collection to the scientific community," says Joanne Kamens, Addgene's Executive Director, "and to promote the great work of Michael Davidson and his colleagues."
Acknowledgments
Thank you to Michael Davidson, Michelle Baird, the Davidson lab scientists, and the FSU team who have made this large, important plasmid deposit possible.
Topics: Fluorescent Proteins, Generating Fusions






Leave a Comment