As optogenetics turns 10 years old, it’s easy to forget that this technique isn’t limited to neuroscience. In fact, precise light-based control of biological processes is highly useful in other fields, including synthetic biology. Addgene depositors Christopher Voigt and Jeffrey Tabor have been working on making E. coli light responsive since 2005, when Tabor was working in Voigt's lab. Years later, these classic systems continue to be optimized by Tabor’s lab, making light-controlled gene expression in E. coli easier and more robust.
Synthetic photobiology: why use light?
In synthetic biology and bioengineering, scientists seek to directly probe and build new systems. Such goals require methods to directly activate and inactivate biological processes. Chemical effectors are suboptimal for a number of reasons, including their potential toxicity and cross-reactivity with other pathways. Since these effectors are diffusible, they’re also not suitable for spatially limited studies and can fluctuate with changing culture conditions.
The use of low intensity light solves each of these problems; inexpensive tools such as LEDs can be engineered to deliver precise, consistent, and controllable pulses of light. Complicated patterns of light intensity and wavelength can be used to study dynamic processes. Conversely, varied input patterns may also be used to engineer new pathways with sophisticated gene expression controls. Most cell types do not respond to low-intensity light, so off-target effects should be limited.
Phytochromes are light-responsive systems found in some bacteria, but not E. coli. They fall under the heading of two-component systems (TCSs). TCSs consist of a histidine kinase that phosphorylates a response regulator (RR). TCS-induced responses are varied; one outcome is directed transcription from a given promoter.
To harness the power of light, Christopher Voigt’s lab created the first E. coli light-sensitive two-component system (TCS). Levskaya et al. fused a photosensory domain from cyanobacteria to a common E. coli histidine kinase. This hybrid construct senses light, and an obligate chromophore allows the system to respond to various light inputs. In far-red light or dark conditions, the chromophore activates the RR (OmpR) via phosphorylation, promoting transcription (see figure below). Subsequent exposure to red light rapidly deactivates the system. Voigt’s lab used this system to develop a bacterial camera that prints a chemical image, as well as a genetic method for the computational problem of edge detection. Tabor et al. subsequently engineered a second photosensitive TCS activated by green light; these two systems can be coexpressed for sophisticated control of gene expression.
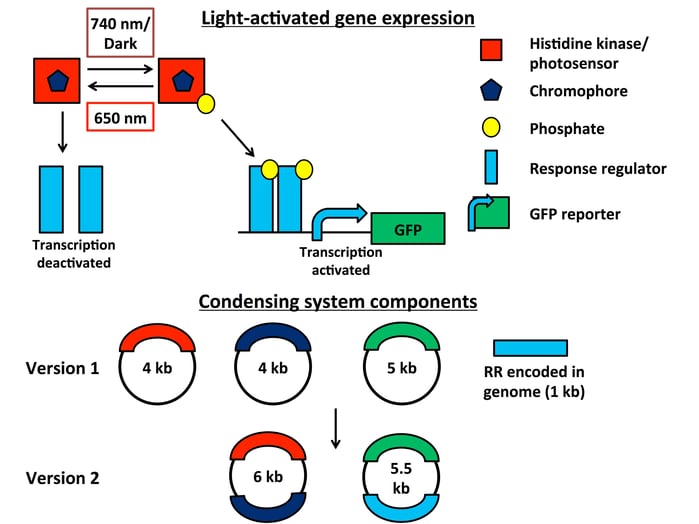
An E. coli light-sensitive two-component system (TCS). A hybrid histidine kinase/photosensor senses various light inputs, and an obligate chromophore allows the system to respond. In far-red light or dark conditions, the chromophore activates the response regulator via phosphorylation, promoting transcription of a GFP reporter. Subsequent exposure to red light rapidly deactivates the system.
Version 1 of this far-red/red light-responsive system is spread across three plasmids, with the response regulator encoded in the genome. To make the system less bulky and easier to use with other strains of E. coli, Schmidl et al. condensed the system to two streamlined plasmids.
The utility of these TCSs was limited by a few factors. These bulky systems were spread over a number of plasmids, with one RR (ompR) encoded chromosomally. Leakiness was also an issue; some promoter activity persisted in the inactive state. The dynamic range, defined as the difference between the lowest and highest level of “output” signal, was only about 10-fold, precluding the study of gene expression at very high or very low levels.
Schmidl et al. chose to optimize these systems to make them more tunable and user-friendly. They experimented with the promoter strength of various components, removing inducible promoters that could crosstalk with other pathways. They also reduced system leakiness and increased dynamic range to around 100-fold. These improvements will allow researchers to better fine-tune the level of output gene expression based on the input light intensity. In addition to these improvements, the constructs have been greatly streamlined; only two plasmids are required per system.
Applications of these systems
The large dynamic range of these systems ensures that they can be used with many types of proteins, including Cas9 and dCas9-transcription factors, for which low expression levels are optimal. These tools are compatible with many strains of E. coli, as well as with other plasmid-based systems, which will enable the assembly of larger systems.
The work of Schmidl et al. also represents a large step forward in optimizing TCSs. Over 75,000 TCSs have been identified in bacteria, and with similar optimization methods, many of these systems could be made suitable for various scientific or engineering applications.
References
Refactoring and optimization of light-switchable Escherichia coli two-component systems. Schmidl SR, Sheth RU, Wu A, Tabor JJ. ACS Synth Biol. 2014 Nov 21;3(11):820-31. doi: 10.1021/sb500273n. Epub 2014 Oct 11. PubMed.
- Find the plasmids from this publication at Addgene.
Synthetic biology: engineering Escherichia coli to see light. Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, Voigt CA. Nature. 2005 Nov 24;438(7067):441-2. PubMed.
- Find the plasmids from this publication at Addgene.
A synthetic genetic edge detection program. Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD. Cell. 2009 Jun 26;137(7):1272-81. doi: 10.1016/j.cell.2009.04.048. PubMed.
- Find the plasmids from this publication at Addgene.
Multichromatic control of gene expression in Escherichia coli. Tabor JJ, Levskaya A, Voigt CA. J Mol Biol. 2011 Jan 14;405(2):315-24. doi: 10.1016/j.jmb.2010.10.038. Epub 2010 Oct 28. PubMed.
- Find the plasmids from this publication at Addgene.
- Check out more synthetic biology posts!
- Check out more optogenetics posts!
Topics: Optogenetics, Synthetic Biology, Plasmids






Leave a Comment