CRISPR technology has changed how scientists edit and control genes, but according to the Broad Institute's Silvana Konermann, the first generation of CRISPR-Cas9 plasmids were not designed with gene activation in mind. “We had not managed to create a system to allow us to reliably activate essentially any gene,” she says. The technical leap from mutating and deactivating a gene or genes to selectively activating them with the CRISPR system was a large one. The question for her then was this: Can you engineer CRISPR-Cas9 activators that work well enough on any gene that they could be used by people with little bioengineering expertise?
“This is the next step in high-throughput, genome-wide screening,” Konermann said. “If you want to activate all 20,000 genes, it is not possible to test each one and make sure it’s working. You can only do that screen with a system you are really confident about working for most genes and individual targeting modules. Those two things, in my opinion, had not been achieved.” That is, until now.
Enter the SAM pooled library
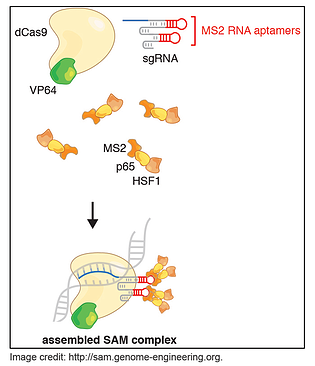 In a paper published in Nature in December, Konermann and her colleagues in Feng Zhang’s lab describe just such a system, which they call CRISPR/Cas9 Synergistic Activation Mediator or SAM. SAM is an engineered protein complex for the transcriptional activation of endogenous genes and it consists of three components:
In a paper published in Nature in December, Konermann and her colleagues in Feng Zhang’s lab describe just such a system, which they call CRISPR/Cas9 Synergistic Activation Mediator or SAM. SAM is an engineered protein complex for the transcriptional activation of endogenous genes and it consists of three components:
1. A nucleolytically inactive Cas9-VP64 fusion;
2. A sgRNA incorporating two MS2 RNA aptamers at the tetraloop and stem-loop 2;
3. The MS2-P65-HSF1 plasmid which expresses the activation helper protein.
SAM can be combined with a human genome-wide library to activate all 23,430 known coding isoforms from the RefSeq database. The key to developing the new system, Konermann explained, was new knowledge about Cas9’s crystal structure.
“I thought maybe we could use information from the structure to guide and improve activator design,” she said. “Before it was a little like tapping in the dark - a lot of trial and error.”
The structural details revealed two exposed RNA loops, which Konermann recognized as an ideal spot to fuse the activators instead of at the protein’s C terminus. With that new design in place, Konermann immediately saw an order of magnitude improvement in activation and knew she was on the right track.
The next insight was to assemble a synthetic transcription activation complex consisting of multiple distinct effector domains that work together synergistically, modeled after natural transcription activation processes. Konermann and her colleagues demonstrated the new system by screening for genes that, when turned on, make melanoma cells resistant to treatment with a BRAF inhibitor. (The test was the perfect complement for one conducted earlier by the Zhang lab for GeCKO, a lentiviral CRISPR library enabling genome-scale, knockout screening).
As the Zhang lab's Nature paper reports, “Here we have shown that the SAM system is robust, specific, and can facilitate genome-scale gain-of-function screening when combined with a compact pooled sgRNA library. Our SAM-mediated screens exhibited a high degree of consistency and validation with >80% effectively enriched guides per gene hit, and 100% validation of the top 10 hits.”
Two-way CRISPR screening
In October, Jonathan Weissman’s lab introduced additional pooled CRISPR libraries, also available at Addgene, enabling systematic investigation of the cellular consequences of repressing or inducing individual transcripts. The activator (CRISPRa) sgRNA library uses the sunCas9 system and contains 10 sgRNAs for each transcription start site in 15,977 human genes. The transcriptional repressor (CRISPRi) library contains 10 sgRNAs for each transcription start site in those 15,977 human genes.
Weissman and his colleagues demonstrated in a paper in Cell that they could typically achieve 90-99% knockdown with minimal off-target effects. The system also enabled them to turn on genes anywhere in the genome for gene control over a 1,000-fold range.
Weissman explained that there are two levels of control in the system. First of all, the Cas9s can be turned on and off, to reversibly activate or repress genes of interest. They also “showed that different guide RNAs have different abilities to turn genes on and off,” he said. “Some are strong and some more moderate; there is an allelic series where the degree to which genes are turned on and off varies depending on which guide RNA we use. What we have to do now is learn more about the rules.”
He said that there is no theoretical limit on the number of genes that can be manipulated at one time. Weissman’s group has made standard protocols available and they are working to further optimize those now. They are also developing sgRNAs that will enable even more fine-tuning of gene expression levels and on software for analyzing the data.
It should be straightforward to apply the new library for use in various cell types and in live mice. Therapeutic uses in humans are also on the table.
“In the future, there is potential there,” he says. "We have to optimize how we can deliver these tools because obviously, unlike the mouse, we can’t rely on genetic engineering to optimize humans for CRISPRi and CRISPRa.”
Before considering the use of pooled libraries in your experiments, Addgene recommends that you review the steps for how to use pooled lentiviral CRISPR libraries and keep the following in mind:
- These libraries are pooled. Most of these screens that can be done with the libraries will require access to next generation sequencing technology.
- These libraries are lentiviral in nature, so please ensure that you are equipped and authorized to make and use lentivirus. Also, note that due to the size of the backbone, transformations require electroporation.
References & Further Reading:
For more information about SAM, including a tool for getting optimized guide sequences for activating any human gene, go to http://sam.genome-engineering.org/.
Crystal structure of Cas9 in complex with guide RNA and target DNA. Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Cell. 2014 Feb 27;156(5):935-49. doi: 10.1016/j.cell.2014.02.001. Epub 2014 Feb 13.
Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Nature. 2015 Jan 29;517(7536):583-8. doi: 10.1038/nature14136. Epub 2014 Dec 10.
Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS. Cell. 2014 Oct 23;159(3):647-61. doi: 10.1016/j.cell.2014.09.029. Epub 2014 Oct 9.
Topics: CRISPR, CRISPR Pooled Libraries







Leave a Comment