Have you ever tried digesting with XbaI or ClaI restriction enzymes and gotten unusual or unexpected results? Or considered why DpnI will degrade your template DNA from a PCR reaction but not the newly synthesized product from a site-directed mutagenesis experiment? The answer to both questions is the same--methylation! Read on to learn about how DNA methylation may affect your restriction digests.

Why methylate?
Restriction enoducleases are so ubiquitous in the lab that it is easy to forget that these enzymes naturally occur in bacteria for purposes other than cloning or confirming plasmids. It turns out that restriction enzymes are one half of naturally occurring restriction modification systems that prokaryotes use to protect themselves from foreign DNA. The other component of these systems, methyltransferases, methylate DNA at particular sequences to prevent them from being degraded by restriction endonucleases. A given prokaryote typically has genes encoding one or a few restriction modification systems containing methyltransferases that add methyl groups to specific DNA sequences and companion endonucleases that recognizes and cleave the same DNA sequence if not methylated (if it comes from a phage for instance).
Aside from restriction modification systems, DNA methylation also plays an integral role in regulating genome replication, repairing mismatched basepairs or small indels that occur during DNA synthesis, and protomoting or repressing protein expression. Methylases involved with these processes (for example Dam and Dcm methylases) are independent from the restriction modification systems, yet can still affect whether certain restriction enzymes can effectively cleave DNA.
Common lab E. coli K12 strains such as DH5alpha contain 3 methylases that recognize and methylate different stretches of DNA:
- Dam methylase adds a methyl group to the adenine of GATC stretches of DNA
- Dcm methylase adds a methyl group to the second cytosine of CCWGG
- EcoKI methylase adds a methyl group to the adenine in AACNNNNNNGTGC or GCACNNNNNNGTT
Impact on cloning and digests
Although not all prokaryotic DNA is methylated to the same level, the potential for methylation should be considered when digesting DNA. Why? Well, even though Dam methylation sites are not specifically associated with any restriction modification systems, their sequences may overlap with restriction sites, inhibiting enzymes such as ClaI or XbaI, or, conversely, activating enzymes such as DpnI.
For example, cleavage by XbaI may be blocked due to methylation if the enzyme's recognition site (TCTAGA) is preceded by GA or followed by TC. As shown in the figure below, a Dam methylase recognition site (underlined in red) overlaps with the XbaI cut site (depicted as an orange line) because the restriction site is followed by TC. Addition of the methyl group at this site would block the enzyme from cutting the DNA there, although other restriction sites (including other XbaI sites not preceded by GA or followed by TC) would be cut as normal. If not taken into account, this blocked site could potentially give you results that would either be difficult to interpret or lead you to conclude your plasmid is not correct!
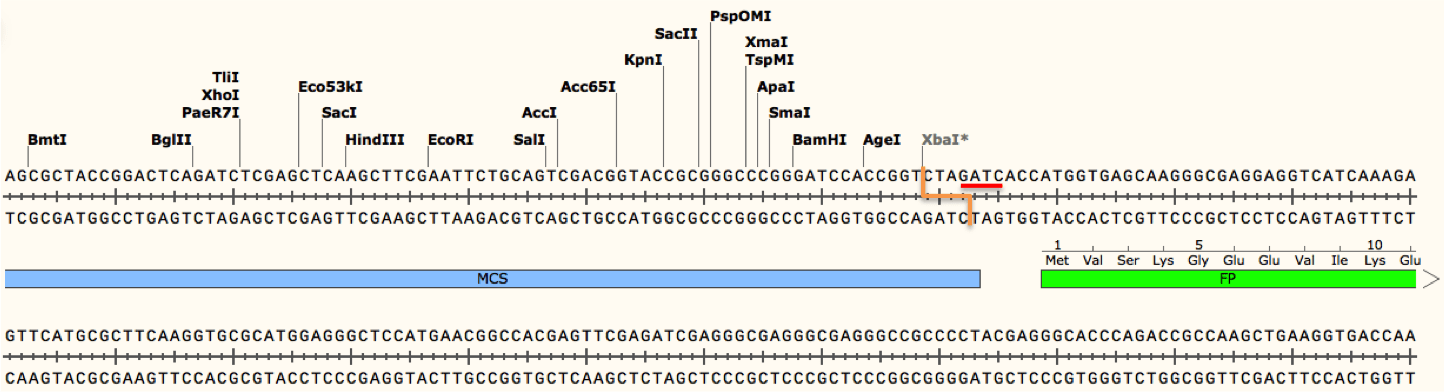
Conversely, enzymes such as DpnI require methylation at their recognition sites in order to efficiently cleave DNA. DpnI is often used for site directed mutagenesis. During this process, incorporation of a desired mutation into your plasmid of interest by PCR generates mutated plasmids with no methylation (there are no methyltransferases in the PCR reaction). The template plasmid, on the other hand, should be derived from a dam+ E. coli strain and will therefore have methylated adenines in any GATC sequences found in the plasmid. When the PCR products are digested with DpnI, only the non-mutated and methylated template is destroyed leaving behind a pool of mutated plasmids which can later be verified by Sanger sequencing.
How can I tell if my enzyme will cut?
Whether performing a digest for cloning purposes or for diagnostics, we suggest double checking to make sure your results will not be affected by methylation. Conveniently, the majority of restriction enzymes commonly used in the lab do not have recognition sites that could overlap with a methylation site. The quick-reference table below lists 10 common enzymes that may be affected by methylation. This table is by no means exhaustive, so you may want to consult the REBASE database for more detailed information.
| Enzyme | Dam methylation | Dcm methylation | EcoKI methylation |
| ApaI | Not affected | Blocked by overlapping methylation | Not affected |
| BsaI | Not affected | Blocked by overlapping methylation | Not affected |
| ClaI | Blocked by overlapping methylation | Not affected | Not affected |
| DraI | Not affected | Not affected | Blocked by overlapping methylation |
| HpaI | Not affected | Not affected | Blocked by overlapping methylation |
| MboI | Blocked by overlapping methylation | Not affected | Not affected |
| MscI | Not affected | Blocked by overlapping methylation | Not affected |
| PmeI | Not affected | Not affected | Blocked by overlapping methylation |
| XbaI | Blocked by overlapping methylation | Not affected | Not affected |
| DpnI | Requires methylation for activity | Not affected | Not affected |
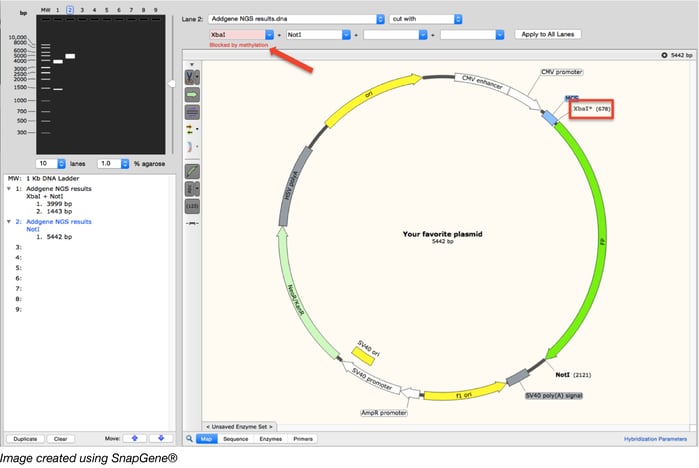
If your enzyme is affected by methylation, knowing the sequence of the DNA surrounding your restriction site(s) can help you quickly predict which (if any) of those sites will be methylated. Using the same example plasmid as above (which has the XbaI site blocked by methylation), the figure below illustrates how sequence visualization software (we used Snapgene here) can help you assess whether your restriction site will be blocked by methylation and how that may change your expected digest results. If Dam methylation is not taken into account, the digest pattern depicted in Lane 1 would be expected for a XbaI/NotI digest. The program, however, allows you to choose whether to look for Dam, Dcm, and/or EcoKI methylation sites and warns you when a chosen enzyme site is blocked. Further, the program takes this into account when simulating a digest as depicted in Lane 2.
Controlling methylation
Finally, you can control methylation by altering your choice of bacteria. For example, if you must use a restriction site that will be blocked by Dam or Dcm methylation, you can ensure this site remains unblocked by first cloning your DNA into a dam–/dcm– strain of E. coli such as JM110 and re-purifying it. These specialized E. coli strains have been specifically engineered to be Dam and Dcm methylase-deficient, and, as such, produce DNA that is unmethylated at those sites. Please keep in mind that dam-/dcm- strains may have an increased rate of mutation (as these would also be deficient in mis-match repair functions of Dam), so these strains should not be used for long term storage.
Additional Resources
- Review of restriction modification systems
- NEB's Resource describing Dam and Dcm Methylases
- Discovery of DpnI
- Review describing dam-/dcm- E. coli
Resources on the Addgene Blog
- Keep up with the Latest Cloning Techniques
- Learn to Troubleshoot Your Sequencing Results
- Use REPLACR Mutagenesis to Mutate Your Plasmids
Resources on Addgene.org
- Check Out Our Plasmid Protocols
- Boost Your Molecular Biology Background
Topics: Plasmids 101, Plasmid Cloning, Plasmids






Leave a Comment