Epigenetics has recently been hitting the headlines, with sotires like the potential devastation of the palm oil industry through epigenetic effects on the Cover of Nature. So what is epigenetics and what tools are available to study it?
What exactly is epigenetics?
Epigenetics is the study of heritable changes in gene expression that do not involve any changes to DNA sequence (Epigenetics literally means ‘on top of the genome’). Epigenetic modifications can result in phenotypic changes without any change in genotype. Epigenetic modifications play a large role in turning genes ‘off’ and ‘on’, signaling which genes should be ‘read’ by the cellular machinery. This is how most cells in the body contain the same DNA but have very different structure and function, and can have a specialized role within an organism.
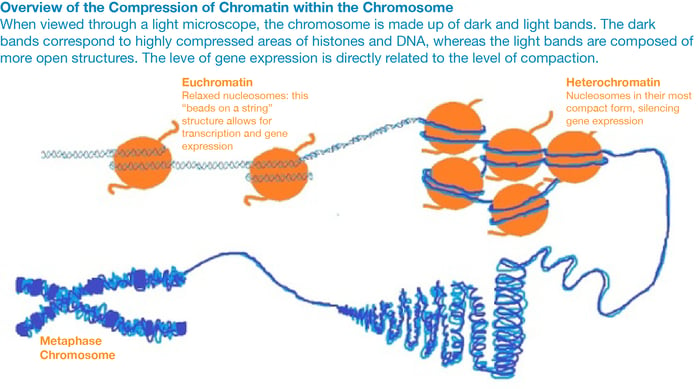
Mechanisms of epigenetic modifications
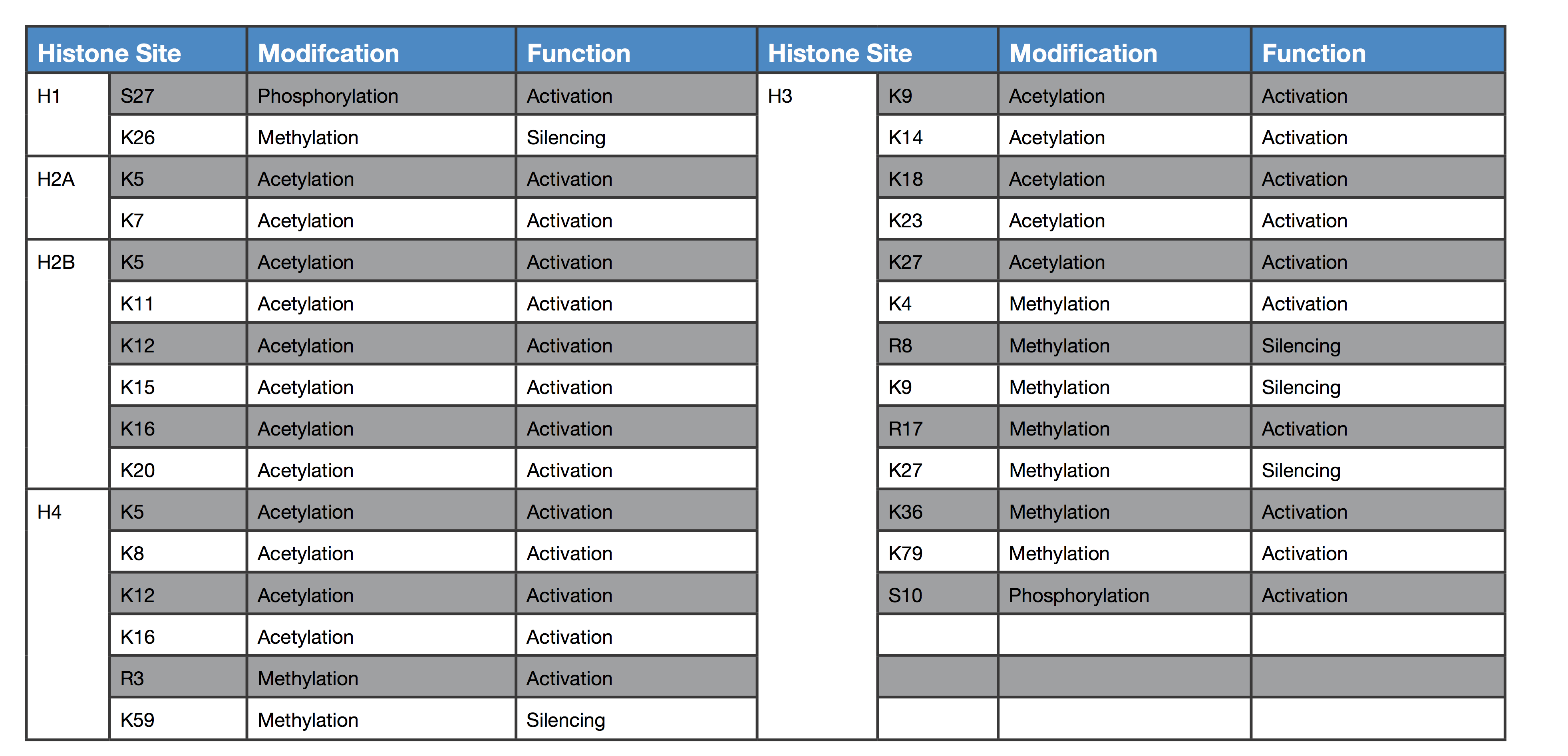
The main mechanism of epigenetic modification is chromatin remodeling. Chromatin is a complex of DNA and histone proteins that compacts cellular DNA into chromosomes and reduces the risk of dangerous DNA damage. If the way DNA is wrapped around histones changes, then gene expression can change as well. The more frequently and tightly bound DNA is to histones, the more compacted it becomes. This prevents the cellular machinery from physically interacting with the genes in the compacted region and results in a loss of gene expression. In this way, genes in compacted regions are essentially ‘silenced’.
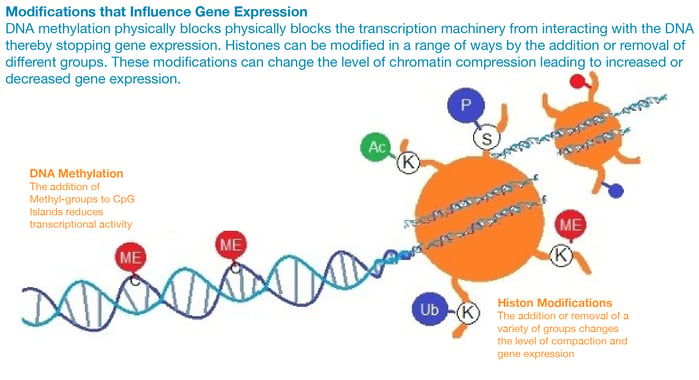
Chromatin remodeling can occur by two main mechanisms, either through histone modifications, which can influence the transcription of genes by RNA polymerase II, or by the addition of methyl- groups to the DNA, which mostly occurs at CpG islands (regions of the DNA with a high frequency of C and G nucleotides). The conversion of cytosine to 5-methylcytosine in CpG islands makes genes less transcriptionally active.
The role of epigenetics in studying disease
Epigenetic changes have been discovered to play a large role in a variety of diseases, ranging from cancer, to diabetes, to congenital disorders.
A series of studies carried out on an isolated population in Sweden found that paternal grandfathers who had experienced famine before their adolescence passed on a heritable trait to their grandsons that decreased their likelihood of death from cardiovascular disease but increased their risk of death from diabetes. Conversely, paternal grandmothers who experienced famine while still in their mother's womb passed on a trait to their granddaughters that reduced their lifespan. Because these dramatic conditions were experienced in a period of life when the grandparents’ gametes were developing, an epigenetic change is thought to have occurred that was passed down to the grandchildren; in this study,epigenetic changes were statistically significantly associated with reduced life-span, but no changes in the DNA sequence could be found.
Angelman syndrome and Prader–Willi syndrome, which have very different disease phenotypes, are two fascinating examples of epigenetics in the form of imprinting. In imprinting, one copy of a gene (either the maternal or paternal copy) is silenced by the addition of methyl groups, and so only the paternal or maternal copy of the gene is expressed in normal individuals (in a parent-of-origin-specific manner), but the expression or silencing of both copies causes disease because too much or too little gene product disrupts normal function.
Angelman syndrome is characterized by intellectual and developmental disability, jerky movements, and spontaneous laughter or smiling, whereas Prader–Willi syndrome is characterized by low muscle tone, short stature, cognitive disabilities, problem behaviors, and a chronic feeling of hunger that can lead to excessive eating and obesity. What is fascinating is that both syndromes can present without any DNA changes, and both involve the same region of chromosome 15. They both result from the inactivation of genes on one copy of chromosome 15 (the maternal copy for Angelman syndrome and the paternal for Prader-Willi syndrome). In both cases, the other copy of the chromosome is normally imprinted and silenced, and so the syndromes are caused by a lack of the corresponding gene products due to the same region, but not the same exact genes, being improperly silenced in both cases. In both diseases, genes from both copies of chromosome 15 are inactivated, but different phenotypes result depending upon whether the improperly imprinted genes come from the mother or father.
Many cancers have dramatic changes in their DNA methylation patterns, either large increases or decreases. Hypermethylation of CpG islands in the promoter region of tumor suppressor genes can repress their transcription and inactivate them. Global hypomethylation, on the other hand, has also been implicated in the development and progression of cancer through different mechanisms.
New FRET based tools to study epigenetics
Epigenetic research uses a wide range of molecular biologic techniques to further the understanding of epigenetics; techniques include chromatin immunoprecipitation, fluorescent in-situ hybridization, methylation-sensitive restriction enzyme digestion, DNA adenine methyltransferase identification, and bisulfite sequencing. However none of these techniques can be used for in-situ research of dynamic modifications in real-time, posing a huge challenge to studying epigenetic changes over time. This challenge was difficult to overcome until now.
Alice Ting’s lab has developed a new, plasmid-based biosensor to visualise changes in histone methylation in living cells. Essentially this reporter construct consists of one substrate (the histone-derived peptide) which can localise the construct specifically to the histone protein of interest (in the figure above, H3 at K9 or K27). Joined to this by a flexible linker is a methyllysine binding domain (chromodomain), which binds selectively to lysine-methylated peptides. This construct is in the middle of a FRET pair, which is used to visualise the results. When methylation occurs at the histone-derived peptide, the methyllysine binding domain causes a structural change (facilitated by the flexible linker) that brings the fluorescent units of the FRET pair together and causes an increase in the FRET signal. Subsequent demethylation separates the fluorescent units and so lowers the FRET signal back again.
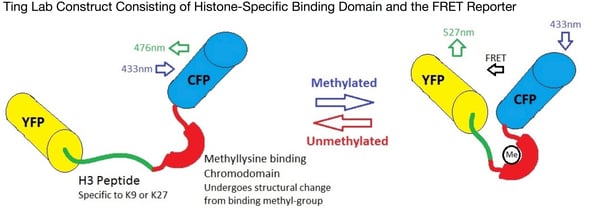
This new technology opens up a new way to visualize changes in gene expression and may lead to the development of a whole new range of methods to study epigenetics. This is likely to have huge impacts in the way we study and treat complex or multifactorial diseases, enabling faster diagnosis and treatment and improving patients lives.
References
1. Ong-Abdullah, Meilina, et al. "Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm." Nature 525.7570 (2015): 533-537. PubMed PMID: 26352475.2. Bygren, Lars O., et al. "Change in paternal grandmothers early food supply influenced cardiovascular mortality of the female grandchildren." BMC genetics15.1 (2014): 12. PubMed PMID: 24552514. PubMed Central PMCID: PMC3929550.
3. Kaati, Gunnar, Lars O. Bygren, and Soren Edvinsson. "Cardiovascular and diabetes mortality determined by nutrition during parents' and grandparents' slow growth period." European Journal of Human Genetics 10.11 (2002): 682-688. PubMed PMID:12404098.
4. Lin, Chi-Wang, Cindy Y. Jao, and Alice Y. Ting. "Genetically encoded fluorescent reporters of histone methylation in living cells." Journal of the American Chemical Society 126.19 (2004): 5982-5983. PubMed PMID: 15137760.Recommended further reading and viewing
- The Epigenetics Revolution: How Modern Biology is Rewriting Our Understanding of Genetics, Disease and Inheritance Amazon
- Ghost in your genes documentary PBS
Additional Resources at Addgene
- Find the Ting Lab plasmids at Addgene
- Browse More Fluorescent Protein Biosensors
- Read Our Tips for Using FRET in Your Experiments Post
- Read Other Blog Posts about Fluorescent Proteins
Topics: Fluorescent Proteins, FRET, Fluorescent Biosensors






Leave a Comment