Updated Mar 26, 2020.
At their most basic level, CRISPR/Cas9 genome editing systems use a non-specific endonuclease (Cas9 or closely related Cpf1) to cut the genome and a small RNA (gRNA) to guide this nuclease to a user-defined cut site. After reading this post, we hope you will be caught up on much of the major CRISPR lingo and will be able to describe the functions of the various CRISPR/Cas9 components. Please note that while this post is intended to provide a general overview of CRISPR components, new Cas9 variants are being discovered all the time and the requirements of these different systems can vary (for example, xCas9 is a variant with increased PAM flexibiliy and eSpCas9/SpCas9-HF1 have increased targeting specificity).
Cas9 endonuclease functions
While native CRISPR/Cas systems have a variety of enzymes responsible for processing foreign DNA as well as the RNA guides required for endonuclease function, when used for genome engineering, the only CRISPR protein required is the Cas9 endonuclease or a variant thereof. This individual protein has all the components necessary to:
1. Bind to a guide RNA
The guide RNA, which will be described in more detail below, enables Cas9 to cut a specific genomic locus of many possible loci. Without binding to the guide RNA, Cas9 cannot cut.
2. Bind to target DNA in the presence of a guide RNA provided that target is upstream (5') of a protospacer adjacent motif (PAM)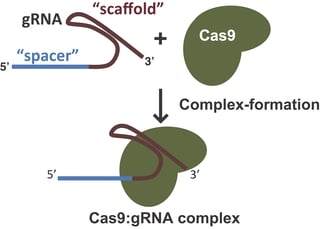
Cas9 endonuclease binding to the target genomic locus is mediated both by the target sequence contained within the guide RNA and a 3-base pair sequence known as the Protospacer Adjacent Motif or PAM. In order for dsDNA to be cut by Cas9, it must contain a PAM sequence immediately downstream (3’) of the site targeted by the guide RNA. In the absence of either the guide RNA or a PAM sequence, Cas9 will neither bind nor cut the target. Cas9 homologs from different organisms or Cas9 mutants developed in a variety of labs (see table below) have different PAM requirements. These different PAM requirements allow researchers to target many different genomic loci.
3. Cleave target DNA resulting in a double-strand break (DSB)
Cas9 and its variants have two endonuclease domains: the n-terminal RuvC-like nuclease domain and the HNH-like nuclease domain near the center of the protein. Upon target binding, Cas9 undergoes a conformational change that positions the nuclease domains to cleave opposite strands of the target DNA. Thus, the end result of Cas9-mediated DNA damage is a DSB within the target DNA ~3-4 nucleotides upstream of the PAM sequence.
Cas9 Species/Variants and PAM Sequences
|
Species/Variant of Cas9 |
PAM Sequence |
|
3' NGG |
|
|
3' NGG (reduced NAG binding) |
|
|
3' NGCG |
|
|
3' NGAG |
|
| 3' NGAN or NGNG | |
| xCas9 | 3' NG, GAA, or GAT |
| SpCas9-NG | 3' NG |
| Staphylococcus aureus (SA); SaCas9 | 3' NNGRRT or NNGRR(N) |
| Acidaminococcus sp. (AsCpf1) and Lachnospiraceae bacterium (LbCpf1) | 5' TTTV |
| AsCpf1 RR variant | 5' TYCV |
| LbCpf1 RR variant | 5' TYCV |
| AsCpf1 RVR variant | 5' TATV |
| Campylobacter jejuni (CJ) | 3' NNNNRYAC |
| Neisseria meningitidis (NM) | 3' NNNNGATT |
|
Streptococcus thermophilus (ST) |
3' NNAGAAW |
|
Treponema denticola (TD) |
3' NAAAAC |
The synthetic guide RNA or gRNA (sometimes sgRNA)
In the native Type II CRISPR/Cas system, Cas9 is guided to its target sites with the aid of two RNAs: the crRNA which defines the genomic target for Cas9, and the tracrRNA which acts as a scaffold linking the crRNA to Cas9 and facilitates processing of mature crRNAs from pre-crRNAs derived from CRISPR arrays. In most systems used for CRISPR-mediated genome editing, these two small RNAs have been condensed into one RNA sequence known as the guide RNA (gRNA) or single guide RNA (sgRNA). Throughout the remainder of this post, we’ll refer to this RNA complex as the “gRNA”. The gRNA contains both the 20 nucleotide target sequence to direct Cas9 to a specific genomic locus and the scaffolding sequence necessary for Cas9 binding. When using CRISPR/Cas9 for genome editing, researchers simply need to express a gRNA designed to direct Cas9 to their target sequence of choice (see tips for designing a gRNA) and their prefered Cas9 variant (with the appropriate PAM sequence) to modify the desired genomic locus.
Some historical notes
CRISPR arrays in bacterial genomes consist of repeated elements separated by unique sequences. When researchers first discovered these arrays, they did not know their functions and simply called the repeated elements “direct repeats” and the unique stretches of DNA between them “spacers” (see figure below).
After years of research, we now know that each direct repeat, combined with its adjacent spacer, ultimately encodes a single crRNA. The direct repeat regions contain sequences required for processing pre-crRNA into mature crRNA and tracrRNA binding. The spacer regions, on the other hand, are the unique, foreign DNA target sequences specific to each individual crRNA.
Bottom line, the “direct repeat region” combined with the tracrRNA forms the scaffold portion of a gRNA and the “spacer region” forms the target sequence.
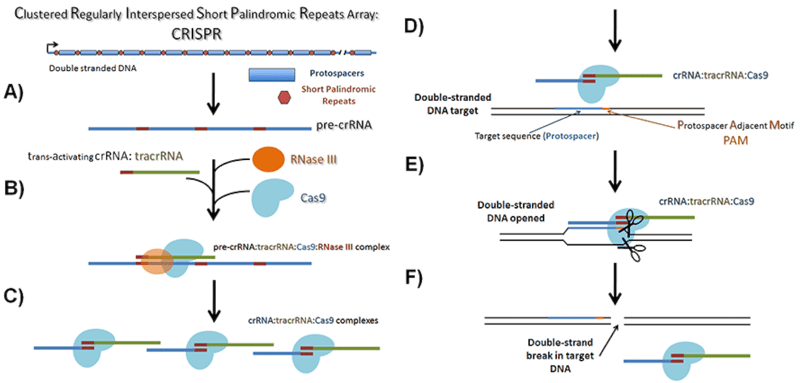 |
|
Overview of native CRISPR arrays and their processing for cleaving foreign DNA. A) CRISPR arrays as found in the bacterial genomes are transcribed into pre-crRNAs containing both the spacer region and the direct repeat region. B) RNase III, the tracrRNA and Cas9, bind to these transcripts and C) cleave them leaving mature crRNAs bound to the Cas9/tracrRNA complex. D) The mature crRNA is used to guide the Cas9 complex to the target DNA which is E) cleaved leaving a F) double-strand break. A “gRNA” is a researcher-designed hybrid of the tracrRNA and the crRNA. The “direct repeat region” combined with the tracrRNA forms the scaffold portion of a gRNA and the “spacer region” forms the target sequence. |
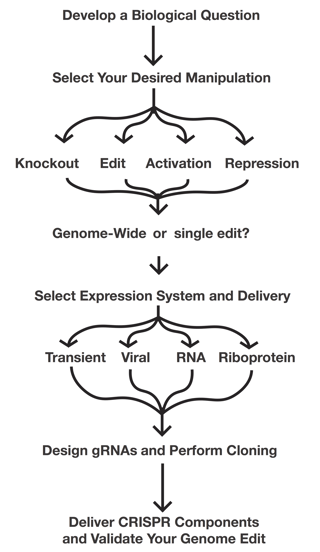
Now that you know about all the different components of CRISPR, It's time to start thinking about how you can used them in your experiments. Use the following flow chart to get you thinking about the types of experiments you're planning on doing and read the eBook or browse our CRISPR resources to flesh-out your plans.
Like what you see? There's much more to be found in the full CRISPR 101 eBook. Happy Reading!
Topics: CRISPR, CRISPR 101







Leave a Comment