This post was contributed by guest blogger Iris Lindberg, Professor at the University of Maryland School of Medicine.
In the Lindberg Lab we often make cell lines that overexpress genes of interest; more recently we have also been using Addgene CRISPR vectors to generate cell lines with knockouts of specific genes. Many years ago, people in the laboratory became frustrated with using glass cloning rings to isolate colonies of antibiotic-resistant cells; during the time required to grease, place and fill a dozen cloning rings, the remainder of the colonies on the plate dried out and died. The alternative to cloning rings, dilution cloning into 96-well plates, is extremely time- and resource-consumptive, since only wells with one cell can give rise to single clones, and thus many plates must be examined for single clones and then handled. Additionally, many cell lines, especially the endocrine cell lines we most commonly work with, require extra serum to survive at low densities - adding to the expense of dilution cloning.
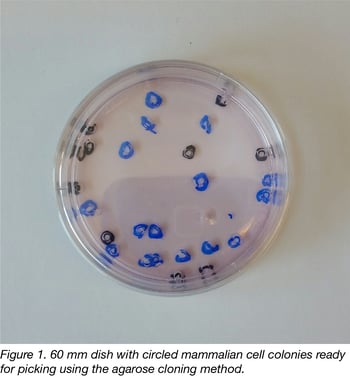
Taking a page from bacterial cloning, we reasoned that we should be able to keep cells moist and hopefully aerated with a thin layer of molecular biology-grade agarose containing physiological concentrations of salts and glucose. If we circled well-separated clones containing more than 50 cells (visible to the naked eye) with a marker prior to agarose addition, we found it was possible to rapidly punch out cell clones with sterile truncated pipet tips. Indeed, an experienced operator can pick 48 clones from a 10 cm dish in 15- 20 minutes- a far more rapid pace than using cloning rings. A picture of a 60 mm dish ready for picking is shown in figure 1.
Find Additional Useful Protocols on Addgene's Protocol Pages
Agarose cloning method procedure
The procedure (illustrated in the YouTube video below) is simple: After obtaining antibiotic-resistant colonies, use a Sharpie to circle 12-48 individual colonies by (carefully) holding plates up to the light, where they appear as opaque dots (colonies should contain at least 50 cells and be well-separated. If the growth rate/transfection efficiency is not known, this is easily achieved by splitting transfected cells into 3 plates at different densities for selection). A 2% agarose solution is then melted (and thereby sterilized) by microwaving and brought into the hood. A 48-well plate is also prepared with about 200 ul of culture medium containing serum in each well. A 200 ul pipettor is set to 200 ul, wiped down with 70% ethanol, and a box of sterile truncated pipet tips (USA Scientific #1011-8410) is also placed in the hood. Finally, a sterile 2X DMEM solution (diluted from 10X and containing both Hepes and bicarbonate) is brought to room temperature and placed in the hood. The medium from a 10 cm dish containing the circled clones is removed by aspiration and the dish left covered (to avoid drying out) while 3 ml of melted agarose are mixed with 3 ml of 2X DMEM in a 50 ml sterile tube (for 60 mm dishes only 1.5 ml of each are used). This solution is capped, swirled and tested for the correct temperature by placing on the wrist or cheek. Within less than a minute, when it is only warm but not hot- and still liquid- it is rapidly poured onto the middle of the plate, where it immediately spreads out to form a firm monolayer. The cell clones - easily identified from the plate circling- are then individually punched out, gently sucked up into the pipettor, placed into wells, and partially dislodged from the agarose plug by rapid pipetting, as shown in the YouTube video. Before returning the plate to the incubator, additional growth medium is added to about halfway up each well.
Tips for using the agarose cloning method
Test your agarose temperature.
Cloning cells using an agarose overlay does have one tricky step. It is necessary to pour the agarose onto the recipient plate at just the right temperature. If the agarose solution is too hot, the cells will die. If the solution is too cold, it will clump prematurely, and not spread out onto the plate. We test the (capped) 1:1 agarose/DMEM solution against a part of the body very sensitive to heat- the inside of the wrist or the cheek. (Parents of infants will have no problem with this!)
Make sure your colonies are well separated.
For this, you need to have some idea of the rate of growth of your specific cell line, and its transfection efficiency. We often trypsinize the transfected cells on the day after transfection into plates containing selection agent at different densities, then dispose of the plates that do not appear to be yielding well-spaced clones. For rapidly growing cells we sometimes use TPP peelaway T-75 flasks to provide a greater surface area (the amount of agarose/DMEM should be proportionately increased).
For some cells, add a drop of serum to aid in outgrowth.
Depending on the cell type, we sometimes add a drop of serum to each well in the 48-well plate; rapidly growing cells such as HEK and CHO do not seem to require this, while nearly all endocrine cells do. While not all of the wells in the 48-well plate will grow up, most will eventually grow within a week. We screen the wells in groups as they become confluent, taking both early- and late-arising clones, and making sure we always maintain a backup replicates of each clone in case of contamination. We do not normally use antibiotics.
Lastly, and perhaps obviously, it is important to have a functional assay in place to screen your clones.
Depending on the protein, we have used Western blots, dot blots, ELISAs, and enzyme assays to select both over- and under-expressing clones. If GFP-encoding cDNA is co-transfected at a favorable ratio to the gene of interest (or, if your protein is fused with a fluorescent protein), fluorescence microscopy can theoretically be used to select specific wells from the 48-well plate for further analysis. To assess CRISPR/Cas9 knockout efficacy, we have used both Western blotting and PCR, depending on the level of endogenous expression.
We highly recommend the use of an electronic pipettor with variable spacing (such as the Viaflo from Integra Biosciences) to transition from a 48-well cell plate to a 96-well assay plate. We generally freeze at least 2 ampules each of 4 clones with different levels of expression of our gene of interest.
Advantages of the agarose cloning method
The agarose technique should be useful to scientists who wish to rapidly isolate individual cell clones with different expression characteristics from a population of transfected clones. The technique is applicable to a wide variety of cell lines, from HEK to CHO cells to neural and endocrine cells such as AtT-20, PC12, Rin5F, Neuro2A and GH4C1, among others (see Lindberg papers in PubMed). While we found that nearly all of the cell lines we tested were amenable to cloning in this manner, we once had difficulty deriving single clones from SH-SY5Y cells, as these cells did not seem to form discrete colonies but constantly migrated away from each other. In our experience, cell clones picked using the agarose method do not differ grossly from those isolated using cloning rings in terms of growth rates, morphology or other physiological properties.
An advantage of this technique is the potentially large number of single clones obtained (on occasion we have prepared two 48-well plates of clones) which enables investigators to identify specific clones with desirable properties, either initially, or using secondary biological screens. For example, for cancer researchers, if growth properties in agarose are of interest, cell clones could potentially be picked following colony formation in selective medium containing a low percentage of agarose, which is then overlaid with the firmer 2% agarose gel solution discussed above for picking.
Many thanks to our guest blogger Iris Lindberg, Professor at the University of Maryland School of Medicine.
 Iris Lindberg is a professor of Neurobiology and Anatomy at the University of Maryland Medical School in Baltimore. She is particularly interested in signaling peptide maturation in neural and endocrine cells. She can be emailed at ilindberg@som.umaryland.edu.
Iris Lindberg is a professor of Neurobiology and Anatomy at the University of Maryland Medical School in Baltimore. She is particularly interested in signaling peptide maturation in neural and endocrine cells. She can be emailed at ilindberg@som.umaryland.edu.
Additional Resources on the Addgene Blog
- Tips for a 1st Time CRISPR User (by a 1st Time CRISPR User)
- The Challenges of Cell Culture
- Plasmids 101: Mammalian Vectors
Resources on Addgene.org
- Check Out Our Protocols Pages
- Browse Fluorescent Protein Plasmids
- Find Mammalian Expression Vectors
Topics: Plasmid Cloning, Molecular Biology Protocols and Tips, Plasmids






Leave a Comment