This post was contributed by Kurt Thorn of the Nikon Imaging Center at UCSF.
A common requirement for live cell imaging experiments is the ability to follow multiple fluorescently tagged species simultaneously. To do so with fluorescent protein labels requires multiple fluorescent proteins whose excitation and emission spectra differ sufficiently for them to be imaged in distinct fluorescent channels on the microscope. With the proliferation of fluorescent proteins in recent years, there are many fluorescent protein combinations that can be imaged together, but this also means that the choice of fluorescent proteins requires some thought.
The first step in choosing fluorescent proteins for your multi-color imaging experiment is to be aware of what fluorescent proteins are available. With new fluorescent proteins being published every month, deciding on the best protein for a given application is a challenge. To help keep you abreast of the latest fluorescent proteins, I maintain an interactive graph and table of the best fluorescent proteins currently available.
Choosing compatible fluorescent proteins
To choose a set of fluorescent proteins to be imaged together, you will need to consider the same factors as when choosing an individual fluorescent protein (brightness, photostability, and so on; see the previous blog post for more discussion of these factors). In addition, you will also need to choose fluorescent proteins that can be distinguished from one another and that can be imaged with the optics on the microscope(s) you intend to use. An accurate determination of whether two fluorescent proteins can be separated from each other requires knowledge of their excitation and emission spectra, but a good rule of thumb is that both the peak excitation wavelengths and peak emission wavelength of the two proteins should be separated by 50-60 nm. For example, CFP (ex 430 nm / em 474 nm) and YFP (ex 514 nm / em 527 nm) can be imaged together but CFP and GFP (ex 488 nm / em 507 nm) show some crosstalk between the two fluorescent proteins. If you must image fluorescent proteins whose spectra overlap, there are techniques, like spectral unmixing, which can be used to separate the fluorescent proteins, but these are beyond the scope of this post.
Are your fluorescent proteins compatible with your microscope optics?
To determine if the fluorescent proteins you are interested in are compatible with your microscope optics, you will want to compare the excitation and emission spectra of your protein with the filter sets or lasers on your microscope. Ideally, you would like to have substantial overlap between the excitation and emission filters and the excitation and emission spectra of the protein, so that the protein is well excited by your microscope and the fluorescence emission of the protein is efficiently collected by the microscope. To compare the match between a fluorescent protein and a filter set, many filter set vendors provide tools to plot the fluorescence spectra of proteins and dyes and their filters (see Chroma's or Semrock's). While these don't contain all fluorescent proteins in common use (particularly not the most recently published ones), they can be a good starting point. In many cases it is sufficient to use a spectrum for a closely related protein, if you know that your protein of interest has a similar spectrum. For example, here's a screenshot from the Chroma Spectra Viewer comparing a standard Cy3 or Rhodamine filter set (Chroma #49004) to the spectra of both mCherry and TagRFP.
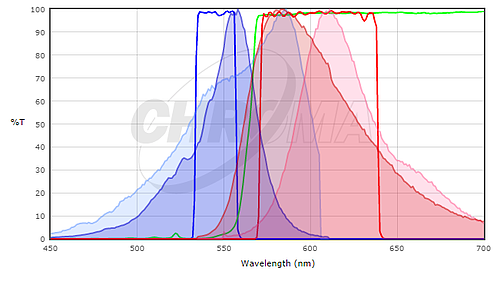
Here, the TagRFP spectrum is shown in the darker colors and the mCherry spectrum is shown in the lighter colors; excitation spectra are blue and emission spectra are red. Neither is a perfect match to the filter set but the excitation filter excites more of the peak of the TagRFP excitation and the emission filter collects a larger fraction of the TagRFP emission than the mCherry emission. For this filter set, we would expect TagRFP to give a brighter signal than mCherry. In general, filter sets designed for Rhodamine / Cy3 will work better with shorter wavelength red fluorescent proteins like TagRFP or mRuby2 than longer wavelength proteins like mCherry. For background on fluorescence and filter sets, see the Introduction to Fluorescence Microscopy lecture at iBiology.
Commonly used filter sets & relevant fluorescent proteins
Commonly used filter sets for multicolor imaging include ones designed for CFP, YFP, and RFP or the Sedat Quad filter set, designed for DAPI / Fluorescein / Rhodamine / Cy5 (e.g. Semrock's) and the similar 4-laser combination on a confocal (405 / 488 / 561 / 640 nm). In our hands the best fluorescent proteins for imaging with this set are mTagBFP2, EGFP or one of the improved GFP variants, mRuby2 or TagRFP-T, and an infrared fluorescent protein such as iFP1.4 or iFP2.0. Beware that these infrared fluorescent proteins require biliverdin as a cofactor and so you may need to supplement your cells with biliverdin for maximal brightness. In mammalian cells, one of the improved folding variants of EGFP like mEmerald or Clover is probably best; mNeonGreen is an even newer green fluorescent protein that is supposed to be extremely bright. In S. cerevisiae, we've tested a number of green and red fluorescent proteins with this filter set and have reported brightness measurements. Here, EGFP outperforms the improved folding variants, presumably due to the lower growth temperature. This also suggests, however, that there is no single fluorescent protein optimal for all organisms and that if you want the brightest signal, you may need to try several proteins in your system of interest. Finally, in this set of proteins the green and red proteins are generally the most detectable and so should be used to tag your least abundant proteins, with the blue and infrared channels used for more abundant proteins or marking compartments.
I hope this sheds some light on multicolor imaging with fluorescent proteins. With the right microscope and the right choice of fluorescent proteins, imaging four colors simultaneously should be pretty straightforward.
Thank You to our Guest Blogger!
 Kurt Thorn is an associate professor at UCSF, where he directs the Nikon Imaging Center. He received his PhD in biophysics from UCSF in the laboratory of Ronald Vale, after which he was a fellow at the Bauer Center for Genomics Research at Harvard University.
Kurt Thorn is an associate professor at UCSF, where he directs the Nikon Imaging Center. He received his PhD in biophysics from UCSF in the laboratory of Ronald Vale, after which he was a fellow at the Bauer Center for Genomics Research at Harvard University.
Topics: Fluorescent Proteins, Fluorescent Imaging







Leave a Comment