Updated Jun 1, 2021 by Meghan Rego.
Reproducible data are key to science, so scientists are used to repeating experiments to confirm their findings. But no scientist wants to repeat an experiment because of poor reagent quality. To make sure our AAV vectors are of the highest quality, we undertake a rigorous quality control process.
In 2016, we launched our viral service to continue our mission of accelerating scientific research. To make sure that adeno-associated viral (AAV) vectors available at Addgene are suitable for in vivo studies, our scientists perform all the standard quality control assays for AAVs. These steps include measuring viral titer and assessing purity. But Addgene is also conducting more rigorous quality control, including next generation sequencing (NGS) of the viral genome to confirm viral genome identity and serotype. Additionally, we check viral transgene expression in transduced cells whenever possible. This post will describe our workflow for viral genome sequencing (VGS).
Why should you care about viral quality control?
AAV vectors are produced by scientists - not robots. Unfortunately, humans have been known to make mistakes. What if a tube is mislabeled or someone grabs the wrong plasmid? What if bacteria snuck into the prep during the production process? These questions are especially important when producing multiple vectors in parallel because the risk of accidental errors increases.
In addition to human error, fragments of DNA from the helper plasmids or the cell genome can be packaged inside the vectors during production. These impurities cannot be removed during the purification step because they are inside the virus itself. While these impurities are generally considered innocuous for research-grade vectors, we’d like to make sure they are present at a very low concentration. So how do we make sure to catch all of these potential mishaps before distributing the vectors?
Viral genome sequencing
Thanks to the establishment of in-house sequencing capabilities and Seqwell’s plexWell technology, we can now perform NGS at Addgene to sequence all the DNA packaged inside the viral particles (Figure 1). Briefly, packaged DNA is extracted from purified, DNase-treated AAV and submitted for NGS. Raw sequencing data is analyzed to determine the identity and serotype of the packaged DNA and look for potential contaminants.
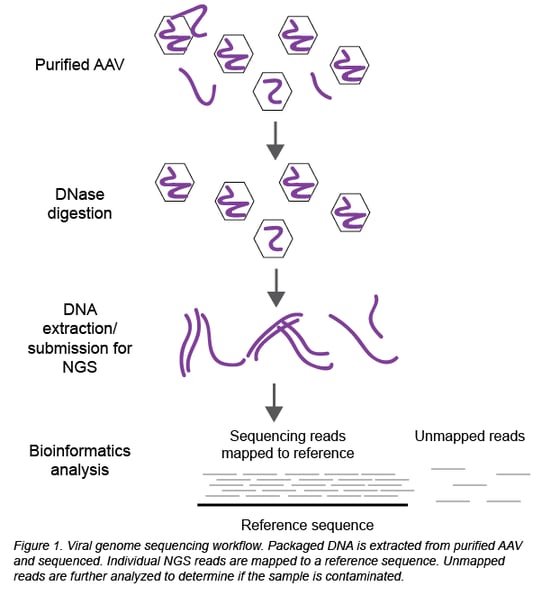
The analysis is a 2-step process using Geneious software:
First, the individual sequencing reads (~150 bp each) are aligned to the reference sequence of the plasmid used to create the viral prep. In a clean viral prep, like AAV-44362 shown below, more than 90% of the reads should align to this reference sequence. Second, we perform a megaBLAST search on all the reads that did not map to the reference sequence. The resulting list of hits is then carefully reviewed. As mentioned earlier, it is common to find DNA from the packaging cell genome, bacterial genomes, cloning vectors, and helper plasmids (Chadeuf et al., 2005, Wright et al., 2011). In fact, the majority of hits come from these known impurities, as was the case for clean sample AAV-44362. In addition to these expected hits, we always get hits to “random” genes. Do these hits always mean the sample is contaminated? No - what really matters is the number of hits to the same sequence (gene). In a clean sample, there are typically fewer than 10 hits to a given sequence, and we believe this isn’t cause for concern. However, a large number of hits to one sequence/gene - we set our threshold at >100 hits - immediately raises a red flag. For example, AAV-68544 contained 220 hits to the CHRM4 gene.
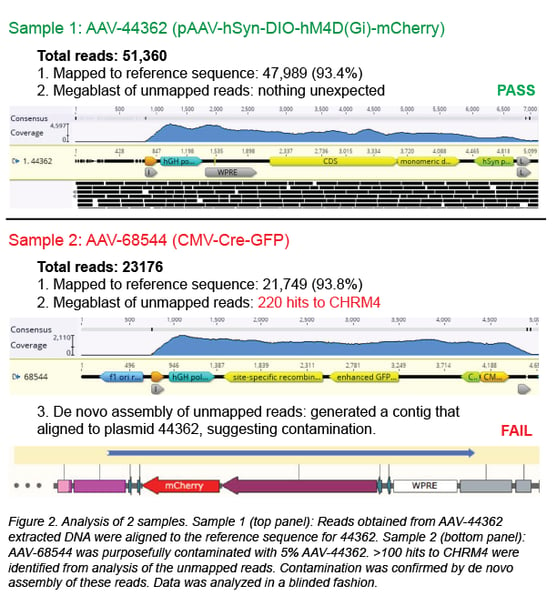
When contamination is suspected, we proceed to de novo assembly of the unmapped reads, employing our bioinformatics software to assemble the reads into longer DNA contigs without using a reference sequence. The five contigs with the most total reads are most likely to correspond to the contaminant. Each contig is then BLASTed against the NCBI nucleotide collection database, and it may also be aligned to plasmids in our inventory containing the suspected contaminating sequence. In most cases, the source of the contamination can be identified, and we will discard any AAV prep we suspect is contaminated. For AAV-68544, the best match for the contaminant is Addgene plasmid #44362.
Method validation, limitations, and future plans
How did AAV-68544 become contaminated? Well, we purposely mixed another AAV vector with AAV-68544 to validate our method. We then extracted the DNA and processed the mixed sample in our pipeline (see Figure 2, sample 2). When we blindly analyzed AAV-68544, we were easily able to identify AAV-44632, the contaminant we mixed into the original AAV sample at 5%. Detecting 5% contamination is a good start, but to determine the sensitivity of our method we wanted to see how low we could go. We prepared a range of spiked samples from 0.1-20% and discovered that, with a high enough sequencing depth, we could detect contamination as low as 0.1% (Guerin et al., 2020).
In addition to genome identification, we developed custom open-access Python scripts that can perform a deeper analysis of the data. Our serotype detection software confirms the serotype of the prep by interrogating the VGS data for small predetermined seed sequences unique to the capsid plasmid. Our recombination software looks for and quantifies the presence of recombinants in recombinase-dependent sequences. Recombinase-dependent sequences may be subject to recombinase-independent recombination during plasmid growth in bacteria or during viral production. This recombination produces a minor pre-recombined population that does not require the recombinase for transgene expression. Once packaged in AAV, this pre-recombined DNA could lead to transgene expression in recombinase-negative animals, producing misleading results. To date, we have analyzed hundreds of viral vectors and estimate that recombinase-independent recombination occurs in 0.1-0.6% of AAV particles in a given viral prep. With this in mind, we recommend that users titrate their AAV vectors to find the optimal dose that allows for sufficient transgene expression but limits the likelihood of recombinase-independent expression.
Next-generation sequencing (NGS) is a powerful tool that can be used to identify DNA contaminants in AAV preparations and provide information on these DNA species with exquisite detail (Lecomte et al., 2015). This higher level of quality control is recommended but not yet required for clinical-grade materials, and is not commonly performed for research-grade materials. Addgene is going the extra mile and pioneering the systematic use of this new QC assay to guarantee that you receive the best AAV vectors for your research.
Please use the comments section below to let us know if you have any specific questions about viral DNA NGS you’d like us to discuss. We are also available by phone or at help@addgene.org to answer your questions about our quality control process!
The images in this blog post were created using Snapgene and Geneious software.
References and resources
References
Chadeuf G, Ciron C, Moullier P, Salvetti A (2005) Evidence for Encapsidation of Prokaryotic Sequences during Recombinant Adeno-Associated Virus Production and Their in Vivo Persistence after Vector Delivery. Molecular Therapy 12:744–753 . https://doi.org/10.1016/j.ymthe.2005.06.003
Guerin K, Rego M, Bourges D, Ersing I, Haery L, Harten DeMaio K, Sanders E, Tasissa M, Kostman M, Tillgren M, Makana Hanley L, Mueller I, Mitsopoulos A, Fan M (2020) A Novel Next-Generation Sequencing and Analysis Platform to Assess the Identity of Recombinant Adeno-Associated Viral Preparations from Viral DNA Extracts. Human Gene Therapy 31:664–678 . https://doi.org/10.1089/hum.2019.277
Lecomte E, Tournaire B, Cogné B, Dupont J-B, Lindenbaum P, Martin-Fontaine M, Broucque F, Robin C, Hebben M, Merten O-W, Blouin V, François A, Redon R, Moullier P, Léger A (2015) Advanced Characterization of DNA Molecules in rAAV Vector Preparations by Single-stranded Virus Next-generation Sequencing. Molecular Therapy - Nucleic Acids 4:e260 . https://doi.org/10.1038/mtna.2015.32
Wright JF, Zelenaia O (2011) Vector Characterization Methods for Quality Control Testing of Recombinant Adeno-Associated Viruses. In: Methods in Molecular Biology. Humana Press, pp 247–278
Additional resources on the Addgene blog
- Check out our Viral Vectors Featured Topic Page
- Subscribe to Viral Vectors Blog Posts
- Learn How We Use NGS for Plasmid Quality Control
Resources on Addgene.org
- Find Ready-to-Use AAV
- Find Ready-to-Use Lentivirus
- Browse Viral Plasmids
Topics: Viral Vectors, Viral Vector Protocols and Tips, AAV






Leave a Comment