If you follow CRISPR research, you know all about using non-homologous end-joining (NHEJ) to make deletions or homology-directed repair (HDR) to create precise genome edits. But have you heard of another double-stranded break repair mechanism: MMEJ (microhomology-mediated end-joining)? MMEJ, a form of alternative end-joining, requires only very small homology regions (5-25 bp) for repair, making it easier to construct targeting vectors. Addgene depositor Takashi Yamamoto’s lab has harnessed MMEJ to create a new method for CRISPR gene knock-in, termed PITCh (Precise Integration into Target Chromosomes). Using their PITCh plasmids, GFP knock-in cell lines can be created in about a month and a half, without the need for complicated cloning of homology arms.
MMEJ: An Introduction
There are three primary methods for repairing DNA after a double-stranded break. HDR copies the sequence from a repair template with flanking sequence homology for error-free DSB repair. NHEJ joins the ends of a DSB in an error-prone fashion, with insertions and deletions common. In contrast, MMEJ uses regions with 5-25 bp of microhomology flanking a DSB to repair DNA. The DNA ends are chewed back to reveal homology, allowing the strands to anneal. DNA synthesis then fills in the gaps. The end result is a deletion of the region between the microhomology and the retention of a single microhomology sequence. For more mechanistic details on HDR and NHEJ, please see the linked blog posts.
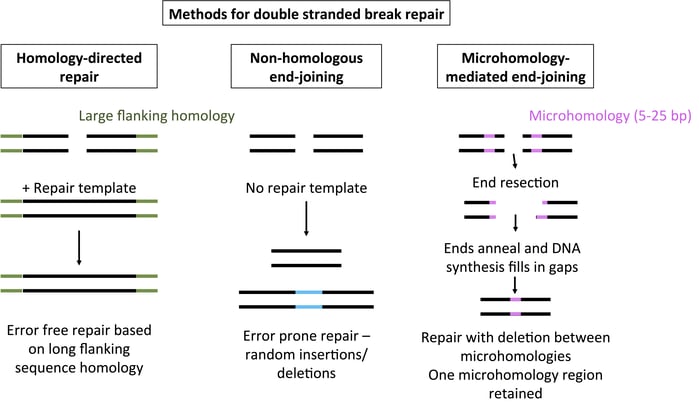
Compared to its counterparts NHEJ and HDR, MMEJ doesn’t get a lot of press. However, this process accounts for a percentage of the mutations seen with TALENs and CRISPR. MMEJ is active during M and early S phases, when HDR is inactive, and the balance of NHEJ, HDR, and MMEJ repair varies from organism to organism. MMEJ doesn’t yield a “perfect repair” like HDR, but it’s much more predictable than NHEJ. As seen in the figure above, short (5-25 bp) regions of homology flanking a double-stranded break yield precise deletion of the sequence between the microhomologies.
MMEJ has certain advantages over HDR. Some species don’t have good HDR systems, and NHEJ will be favored even if a repair template is present. HDR also presents a cloning dilemma - the longer the homology, the more efficient the recombination, but with longer homology arms comes more time spent cloning. In contrast, the short homology required by MMEJ can easily be added via PCR amplification. Given the inefficiency of HDR for knock-ins, some labs have used NHEJ for whole plasmid integration; however, since NHEJ is error-prone, such a system is likely to introduce additional nucleotides flanking the insertion. If the DNA ends anneal incorrectly, MMEJ may also introduce, substitute, or delete nucleotides in addition to the expected deletion, but the frequency should be lower than that observed with NHEJ.
PITCh: Using MMEJ for gene knock-in
Building on the lab’s previous work, Sakuma et al. describe a detailed protocol for MMEJ-mediated knock-in of a GFP-Puro cassette into a given locus, just upstream of a stop codon. Briefly, the PITCh vector should be constructed with 5’ and 3’ microhomology to the insertion locus flanking the GFP-Puro cassette. Three double stranded breaks are necessary for knock-in: one on either side of the GFP-Puro cassette and one in between the 5’ and 3’ microhomologies in the genomic locus. The first two breaks are induced via a generic PITCh-gRNA; the third break by an insertion locus-specific gRNA. These double stranded breaks allow for two sets of microhomologies (5’ and 3’) to anneal, knocking the GFP-Puro cassette into the locus (see figure below). The double-MMEJ strategy looks very similar to HDR, but it is accomplished using much smaller regions of homology, which facilitates easier cloning.
Abbreviated protocol for PITCh
Step 1: Generate microhomologies in the PITCh vector
~20 bp 5’ and 3’ microhomologies are added to the GFP-Puro cassette via PCR, and this construct is inserted into the PITCh vector via In-Fusion or SLIC cloning.
Step 2: Design an insertion locus-specific gRNA
The gRNA should target near the last coding exon of your gene of interest. For ideal use, this gRNA should be cloned into a vector containing Cas9 and the PITCh-gRNA.
Step 3: Contransfect the PITCh vector with the vector carrying Cas9 and the PITCh- and locus-specific gRNAs
Step 4: Select for puromycin resistant cells
Step 5: PCR amplify and sequence the locus to verify correct GFP-puro insertion
To lower the risk of off-target effects, Sakuma et al. optimized the PITCh-gRNA for minimal off-target binding in mammalian genomes as assessed by CRISPR design tools at crispr.mit.edu. They tested their system in HEK293 cells, integrating the GFP-Puro cassette into the FBL locus. Upon sequencing of puromycin resistant clones, they found that 80% and 50% of clones displayed proper insertion at the 5’ and 3’ junctions, respectively. These results indicate that PITCh is a robust method for genomic insertion. PITCh can also be adapted for whole-plasmid integration if you’d like to integrate a larger amount of material into the genome.
Open questions and alternative systems
The ready-made PITCh plasmids available from Addgene are perfect for expressing GFP from a given promoter, and the technique can be adapted to other transgenes. It’s important to note that the fluorescence level observed will be dependent on both the endogenous promoter and the 3’ UTR of the locus of interest, since the GFP-Puro will be inserted just upstream of a stop codon. One potential concern is if the GFP-Puro will alter expression of the gene it follows.
For increased versatility, it would be advantageous to adapt PITCh to insert genes into AAVS1, the “safe harbor locus” of the human genome, as shown by Dalvai et al., who used HDR to insert cDNA constructs into this locus. One important question to ask is how the efficiency of PITCh-based genomic insertion would compare to CRISPR sticky-end insertion using the nuclease Cpf1. Since Cpf1 cuts in a staggered pattern, it is thought to be ideal for HDR-independent knock-ins, but this possibility is still being explored.
More broadly, Sakuma et al.’s use of MMEJ represents another strategy researchers can exploit for CRISPR gene editing. In organisms where HDR is downregulated, MMEJ represents another method for making targeted modifications. A recent publication by Zhang et al. shows just that - using MMEJ to insert FLAG tags into the genome of the pathogenic fungus Aspergillus fumigatus, which has been difficult to modify due to NHEJ’s dominance over HDR in this species. As CRISPR technology continues to develop, it’s become clear that the power of this editing platform lies in the diversity of nucleases and their applications. It will be interesting to see what new editing possibilities MMEJ can enable.
References
1. Sakuma, Tatsushi, et al. (2016). “MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems.” Nat Protoc. 11(1): 118-33. PMID: 26678082.
- Find the plasmids from this publication at Addgene.
- Find plasmids from this publication at Addgene.
3. Zhang, Chi, Xiuhua Meng, Xiaolei Wei, and Ling Lu. (2016). “Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus.” Fungal Genet Biol. 86: 47-57. PMID: 26701308.
Additional Resources on the Addgene Blog
- Brush up on Homology Directed Repair and Non-homologous End Joining
- Read About Other Genome Engineering Tools like Sleeping Beauty Mutase
- Find the Ideal CRISPR Software for Your Experiment
Additional Resources on Addgene.org
- Learn All about CRISPR on Our CRISPR Guide Pages
- Find CRISPR Plasmids for Your Experiments
- Deposit Your New Plasmid Tools with Us
Topics: CRISPR, CRISPR Protocols and Tips, Other CRISPR Tools








Leave a Comment