Laboratory management software is not a requisite for a functioning lab, but it is for a scaleable lab. When you need to track the location, quality, growth, and legal status of thousands of plasmids a day, like we do at Addgene, pen and paper will fail you. The benefits of lab management software aren’t just limited to large volume facilities; it can be useful in academic labs where postdocs, students, and lab mates are coming and going frequently - an environment ripe for valuable work and materials to slip through the cracks.
In this post we’ll highlight some of the lessons we’ve learned over the years with the hope that our insights can help steer you in the right direction when writing your own software. None of this is gospel, but we think it's worth consideration.Everything will go wrong
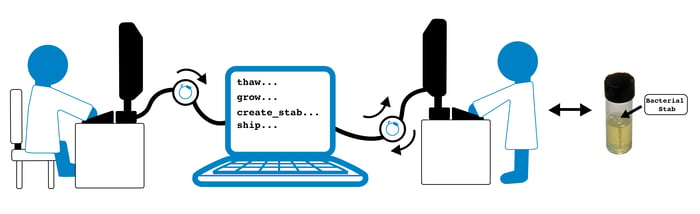
 Whenever an order is placed on the Addgene website, it sets off a chain reaction of events throughout all of Addgene - within the office, within our legal department, and, of course, within the lab. In order to get a plasmid to your lab bench, we need to (this is a little simplified):
Whenever an order is placed on the Addgene website, it sets off a chain reaction of events throughout all of Addgene - within the office, within our legal department, and, of course, within the lab. In order to get a plasmid to your lab bench, we need to (this is a little simplified):
- Find the appropriate glycerol stock within our freezers
- Pick the sample into the appropriate stab culture
- Grow the stab culture
- Ship the stab culture to the proper location
To keep ourselves from mixing up samples and to ensure that everything is grown successfully, we must track and manage all of these steps. Unfortunately, biology is messy. Every single step in a lab procedure can and will fail. Some step failures require re-attempting the same step, some require going back two steps, some require starting over entirely, and our software needs to handle these cases. The following code examples show how software can be structured to allow for the messiness of biology. These examples are intentionally trivial, but they're here to help tell this story.
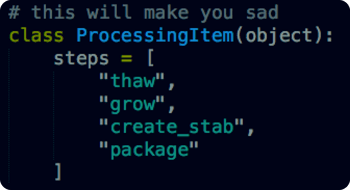 In this first example to the right (# this will make you sad), someone has put in a request for a particular plasmid and now it’s time for our lab technicians to retrieve the sample and get it ready for shipping. In the first case, we naively assume that everything will go perfectly and our software simply follows a linear progression directing our techs through the process.
In this first example to the right (# this will make you sad), someone has put in a request for a particular plasmid and now it’s time for our lab technicians to retrieve the sample and get it ready for shipping. In the first case, we naively assume that everything will go perfectly and our software simply follows a linear progression directing our techs through the process.
The problem? As we said above, any one of these steps can fail but, in our current setup, there are no explicit instructions on what to do when something fails. While we’re sure our techs could figure it out if they needed to, they’d have to individually keep track of any failures and do troubleshooting on their own. For a few samples this isn’t so bad, for thousands it’s monumentally difficult without software.
Tracking lab procedures also gives you access to hard data about what steps are most likely to fail. Our laboratory manager would definitely be interested to see which part of her procedures are the most failure prone.
In an academic lab, a similar situation could arise if a lab veteran who worked with a particular sample suddenly left and didn’t have a system through which she could easily transfer her instructions for dealing with the samples to newer lab members.
We can set ourselves up for success by giving explicit instructions to lab members through the software if there is either success or failure:
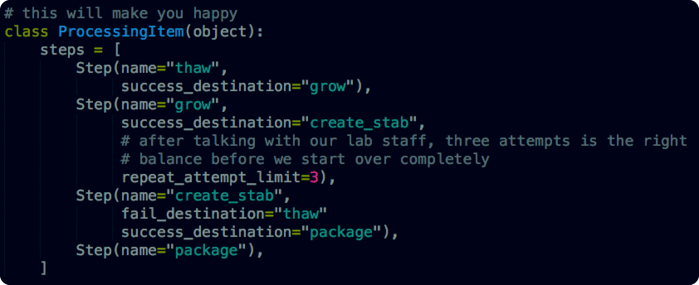
In our second example we’ve added some simple but critical logic. Some steps fail “hard”, they reset the whole procedure. Other steps fail “soft” they simply need to be repeated. We also now have a good interface to begin tracking analytics data for lab procedures. With this data in hand, we can go back and optimize our procedure to make the whole process work more efficiently.
Get on that barcode gravy train
- Buy a barcode label printer.
- Buy barcode scanners.
- Barcode everything.
- If it’s barcoded, it should have a corresponding entry in your database.
- Enjoy your happy lab staff.
Barcoded materials make everyone's life easier, the time and capital cost of getting up to speed with a setup is almost always worth it. Because labels are applied to containers and not the mushy, gooey contents of said containers, think carefully about how you want your software to treat those encoded numbers.
Loosely couple containers and contents
Right from the get-go be as clear as you can be (there will always be confusion about this though) about the fact that containers are distinct from their contents. You should be tracking both, you should easily be able to move contents from one container to another.
If you are using barcodes (and you should be!) bear in mind that your barcodes are being applied to the containers, not to the contents. Tightly coupling barcodes and containers might not be such a bad idea, tightly coupling them to their contents might leave you sad and confused when a tricky procedure failed and one of your techs started over with a new tube.
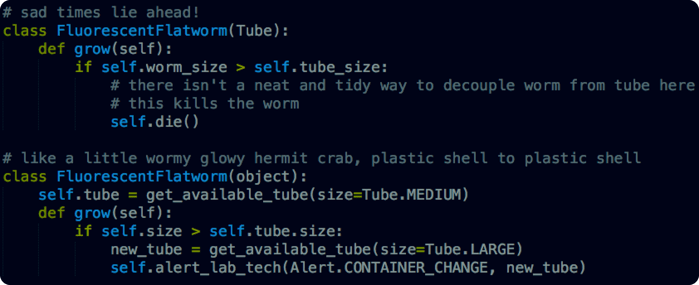
In the first example we’ve used inheritance to tightly couple the contents of a tube with the tube itself. If something happens to the worm that requires a change of tube, there’s no clean way to separate the contents from the container (of course this is a simple task in the lab, but the software should be mirroring the lab procedures, not artificially constraining them).
In the second example we have a separate worm model and tube model, we can interchange them easily as needed. In this case, the physical considerations of the laboratory make object composition a smarter choice than inheritance.
Some containers are permanent, like a tray or a freezer. Some are disposable, like a plastic tube. Talk with your lab team about how they would like these things tracked.
Terminology and language are tricky
Names for things will trip everyone up at some point, decide early on what to do in a namespace collision. It may even be worth it to have extra columns in your database to the effect of `dev_name` and `science_name`. You'll need to decide given the technical details of the problem you're solving, but consistency is key. Some namespace collisions you may encounter: vector, sequence, insert, trait, factor, expression, etc.
There are also going to be situations where you learn biology terms (plasmid, 5-prime, CRISPR, growth strains, vector backbone, etc.) and you simply don't have enough context to know what they really are. Your code could be treating these pieces of data in ways that are inconsistent with how they are being treated in the lab, because of this, it is important to write loosely coupled, modular code.
Use tried-and-true tools
We use Python, Django, Apache, and MariaDB. Using tried-and-true tools means less time getting up and running and more time working on interesting biotechnology code.
There are valid technical reasons to choose a non-Python stack, but if you're working with scientists it's unlikely they will have exposure to anything other than Python or MATLAB. Choosing Python makes it easy to spin up a read-only Jupyter notebook for them and let them in on the fun too.
 Kris Shamloo is a Software Engineer at Addgene and is interested in writing software tools to help scientists. You can find him on Twitter @krisshamloo and at krisshamloo.com
Kris Shamloo is a Software Engineer at Addgene and is interested in writing software tools to help scientists. You can find him on Twitter @krisshamloo and at krisshamloo.com
Additional Resources on the Addgene Blog
- Learn about How We Use Barcodes in the Lab
- Get Tips on How to Keep a Lab Notebook for Bioinformatic Analyses
- Use Software Apps to Save Time and Track Your Experiments
Additional Resources on Addgene.org
Topics: Addgene News






Leave a Comment