Originally published Jan 28, 2016 and last updated Sep 10, 2020 by Jennifer Tsang.
CRISPR makes it easy to target multiple loci - a concept called multiplexing. Since CRISPR is such a robust system, editing or labeling efficiency doesn’t usually change when you add multiple gRNAs on one plasmid. Sound good? Addgene has many tools to help you multiplex - we’ll use mammalian plasmids to introduce you to some of your potential options and cloning methods, but please scroll down for plasmids suitable for other model systems, including E. coli, plants, Drosophila, and zebrafish!
Why use multiplexed gRNAs?
By expressing multiple gRNAs on the same plasmid, you’ll make sure that each cell that gets the plasmid contains all of the desired gRNAs. This increases the chance that all the edits you want to make will happen.
There are many reasons to multiplex gRNAs, some being:
- Using dual nickases to generate a knockout or edit. This can help reduce off-target activity.
- Deleting a large region of the genome by removing the sequence between two target sites.
- Modifying multiple genes at once. Using multiplexed gRNAs can target multiple locations in the genome to modify, whether it be editing CRISPRi, CRISPRa, or base editing.
Multiplexing gRNAs: the basics
One common question Addgene Senior Scientists receive is: can I express more than one gRNA from a single promoter using a plasmid like pX330? Unfortunately, the short answer is no. Unless you use a system for processing a continuous multi-gRNA transcript, each gRNA must be expressed from its own promoter. But that doesn’t mean you have to clone and transfect multiple promoter-gRNA constructs in order to target multiple sites! In this post, we'll cover Cas9 multiplexing options, but also check out our blog post about multiplexing with Cpf1.
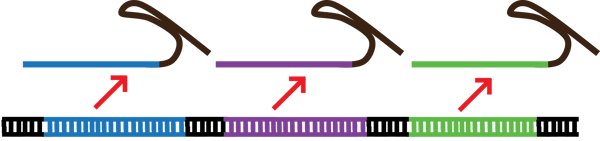 |
| Figure 1: Multiplexing allows researchers to express multiple gRNA from a single construct. DNA and gRNA are not to scale. |
Let’s start with the simplest multiplexing situation: you only need to express two gRNAs at the same time. One system you could use is pX333 from the Ventura lab. pX333, a modification of pX330, contains humanized wtCas9 and two U6 promoters. To use this plasmid, you simply order oligonucleotides for your chosen gRNA target sequences and clone them in just as you would for a single gRNA. You’ll clone in the first gRNA using restriction enzyme BbsI and the second gRNA using restriction enzyme BsaI. If you’re working in Drosophila, a two-gRNA expressing plasmid is available from the Bullock lab, and gRNAs can be inserted using Gibson Assembly or SLIC cloning methods. A BsaI-based E. coli multiplexing plasmid is available from the Koffas lab.
Scaling up your multiplexed gRNAs with Golden Gate and Gibson assembly methods
If you want to scale up the number of gRNAs in your plasmid, you’ll need to use some assembly methods such as Golden Gate or Gibson assembly.
Golden Gate Assembly of gRNAs
Golden Gate Assembly uses Type IIS restriction enzymes, which cleave outside of their recognition sequence, creating flanking overhangs. These overhangs can be customized to link together multiple fragments, allowing ordered assembly of multiple components into a destination vector.
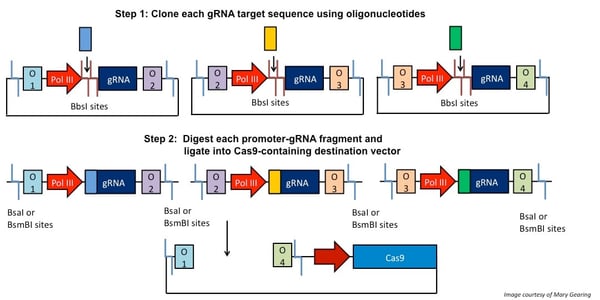
The first step in CRISPR/Cas9 Golden Gate multiplexing is to clone the oligonucleotides specifying each gRNA target sequence into distinct expression vectors using the enzyme BbsI. These expression vectors each contain Type IIS restriction sites flanking the promoter-gRNA construct, but with different sequences adjacent to the sites. When digested with the appropriate Type IIS enzyme, the unique flanking overhang sequences can link together to allow for ordered assembly into a destination vector that expresses Cas9. This is illustrated in the schematic below.
|
|
|
Figure 2: gRNA target sequences (colored rectangles) are cloned into various plasmids using oligonucleotides. These plasmids contain Type IIS restriction sites that flank the promoter-gRNA constructs. When these plasmids are digested, unique overhangs (here, O1-4) adjacent to the cut sites “link” fragments together and drive ordered assembly into a Cas9-containing destination vector. Note: depending on which method you use, the procedure will vary slightly. |
Gersbach Lab multiplexing plasmids:
This plasmid set allows you to express 2-4 gRNAs, with four being the ideal number. First you generate four unique kanamycin-resistant plasmids, each containing a different gRNA target sequence downstream of the 7SK, human U6, mouse U6, or human H1 promoters. If you express fewer than four gRNAs, you’ll clone in a polyT-termination sequence for each unused promoter. This step is necessary to generate all of the overhangs needed for the final ligation step. Plasmids are then digested using BsmBI and ligated into Cas9 or dCas9-containing destination vectors. Destination vector options include humanized wt Cas9, dCas9 (transcriptional repressor), and dCas9-VP64 (transcriptional activator)-containing plasmids. Each destination vector contains GFP, enabling you to select cells with high GFP expression. These cells have the highest levels of Cas9 and gRNA expression, and thus the highest frequency of genome editing events.
Yamamoto Lab Multiplex CRISPR/Cas9 Assembly Kit:
This kit is built for serious multiplexing and enables users to express up to 7 gRNAs. The kit contains different destination vectors depending on the total number of gRNAs you wish to clone, from 2-7. For example, if you’re expressing 4 gRNAs, you’d use pX330A-1x4; for 6 gRNAs, you’d use pX330A-1x6. This customization means you don’t ever need to clone in filler sequences. To build your multiplexing construct, you clone all but one of your gRNAs into spectinomycin-resistant plasmids pX330S-2 to pX330S-(last gRNA number). The 5’ most gRNA is cloned into the Cas9-containing destination vector. These constructs are digested using BsaI and assembled to produce a plasmid encoding the gRNAs and Cas9. As with the Gersbach lab plasmids, multiple Cas9 variants are available: wt humanized Cas9, D10A nickase mutant (Cas9n), dCas9 (transcriptional repression), and Fok1-dCas9 (dimeric nuclease).
Gateway assembly method
Frew Lab Multiple Lentiviral Expression Systems (MuLE) Kit:
This kit can be used to create lentiviral vectors expressing wt humanized Cas9 and up to three gRNAs. Entry vectors containing the U6 promoter and the gRNA scaffold are provided with the kit. Oligonucleotides specifying the gRNA seed sequence should be compatible with type IIS enzyme BfuAI. Gateway cloning is then used to combine the multiple gRNAs and Cas9 together into a single plasmid. Although only wt hCas9 entry vectors are supplied with the kit, you can clone your own entry vectors containing other Cas9 variants to use with the MuLE system.
Multiplexing from a single transcript
You can also multiplex gRNAs via a polycistronic transcript. Rather than being transcribed from different promoters, the gRNAs are transcribed together and are flanked by specific sites that allow them to be cleaved and released. These constructs tend to be smaller than constructs with multiple promoter-gRNA cassettes, making them advantageous for small capacity vectors like AAV. In addition to the mammalian option described below, plasmids for making polycistronic gRNAs are also available from the Yang lab for use in plants.
The mammalian multiplex systems use the Csy4 RNA nuclease from Pseudomonas aeruginosa. When overexpressed, Csy4 efficiently cleaves gRNAs sandwiched between 28 base Csy4 recognition sites. If Csy4 is not expressed, the gRNAs cannot be released, adding temporal and/or spatial control to the system. pSQT1313 from the Joung lab allows you to express two gRNAs constructed using oligonucleotide assembly. Unlike some of the plasmids described above, this vector does not contain Cas9, so you’ll need to supply it with another plasmid.
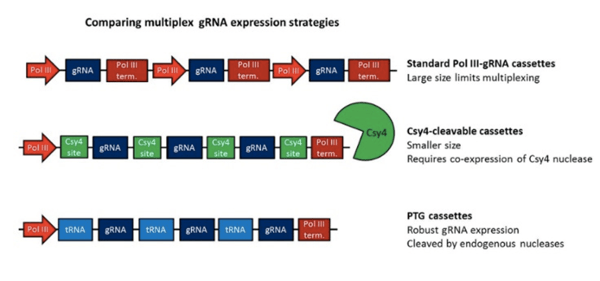 |
| Figure 3: Comparison of Multiplex Strategies including Standard PolIII-gRNA cassette, Csy4-cleavable cassette, and PTG cassette. |
Find gRNA multiplexing vectors at Addgene!
Multiplexing in plants
Qi-Jun Chen Lab Golden Gate/Gibson Assembly Multiplexing Plasmids:
 These plasmids allow you to assemble 2-4 gRNAs through Golden Gate or Gibson Assembly. gRNAs are inserted into the pCBC vectors using BsaI, and promoter-gRNA fragments are PCR amplified for cloning into one of three Zeamays codon-optimized Cas9-containing binary vectors. The system is compatible with both monocot and dicot plants.
These plasmids allow you to assemble 2-4 gRNAs through Golden Gate or Gibson Assembly. gRNAs are inserted into the pCBC vectors using BsaI, and promoter-gRNA fragments are PCR amplified for cloning into one of three Zeamays codon-optimized Cas9-containing binary vectors. The system is compatible with both monocot and dicot plants.
Liu Lab Golden Gate/Gibson Assembly Multiplexing Plasmids:
These plasmids can be used to express up to 8 gRNAs after Golden Gate or Gibson Assembly. Using BsaI, gRNAs are cloned into one of 12 pYLsGRNA plasmids, which contain various promoters and reporters, and subsequently inserted into a Cas9-containing destination vector based on pCAMBIA. Cas9 is plant-optimized, and plasmids are available for both monocot and dicot plants, with a choice of either hygromycin or Basta selection.
Yang Lab Single Transcript Multiplexing Plasmids:
These plasmids are similar to the Csy4 polycistronic system described above, except that they use an endogenous nuclease system to cleave the gRNAs. gRNAs are flanked by glycine tRNAs to create polycistronic glycine tRNA-gRNA (PTG) constructs. Eukaryotic RNases P and Z recognize the tRNA sequences, cleave them, and release the gRNAs. PTGs are assembled into a wt Cas9-containing vector using Golden Gate assembly, and up to 8 gRNAs may be expressed simultaneously. Vectors for both transient expression and Agrobacterium-mediated transformation are available. For more information on this system, check out this blog post.
Multiplexing in zebrafish
 Wenbiao Chen Lab Golden Gate Assembly Multiplex Plasmids:
Wenbiao Chen Lab Golden Gate Assembly Multiplex Plasmids:
These plasmids allow expression of 2-5 gRNAs in zebrafish. Custom destination vectors are used depending upon the total number of gRNAs you wish to clone, so you don’t have to clone any filler sequences. You also have the option of including a previously-designed tyr gRNA, which causes hypopigmentation, thus marking cells that have undergone genomic modification. In this system, Cas9 must be supplied on a separate plasmid.
Multiplexing in Drosophila
 Bullock Lab Multiplex Plasmid:
Bullock Lab Multiplex Plasmid:
Two gRNAs can be assembled using Gibson Assembly or SLIC cloning. gRNAs are expressed from two Drosophila U6 promoters. Cas9 must be supplied on a separate plasmid.
Multiplexing in E. coli
 Koffas Lab CRISPathBrick Multiplex Plasmid:
Koffas Lab CRISPathBrick Multiplex Plasmid:
This system allows you to assemble type II-A CRISPR arrays for dCas9-based transcriptional repression. The CRISPathBrick plasmid contains a nontargeting spacer flanked by two CRISPR repeats. The spacer can be digested using BsaI, allowing a spacer-repeat “brick” to be inserted. The BsaI site remains intact, allowing subsequent “bricks” to be added one by one. This approach is especially useful for combinatorial analyses. For example, if you were to develop an array using 3 distinct spacer-repeats, you could easily create 7 unique arrays (e.g. for spacers A, B, and C, you could obtain arrays A, B, C, AB, AC, BC, and ABC).
Nielsen Lab E. coli CRMAGE Plasmids:
The CRMAGE system is a fast, multiplexable method that combines CRISPR and recombineering-based MAGE (Multiplex Automated Genome Engineering) technology. pMA7CR_2.0 expresses lambda Red and Cas9, which are separately inducible by L-arabinose and anhydrotetracycline (aTet), respectively. pMAZ-SK contains an aTet-inducible gRNA and a backbone-targeting gRNA cassette for plasmid curing through "self-destruction" after induction with L-rhamnose and aTet. CRMAGE is much more efficient than traditional recombineering, with 96-99% efficiency for point mutations and 66% efficiency for small insertions. Multiplexing of two targets simultaneously is possible with efficiency >70%. CRMAGE is an incredibly fast protocol, with only 5 hours incubation time needed for a single round of editing, and a subsequent curing protocol that requires only 2-3 hours incubation.
Kondo Lab multiplexed base editing in E. coli
The Kondo lab expressed a cytidine deaminase fusion with Cas9 with a uracil DNA glycosylase inhibitor with a degradation tag (LVA tag) to achieve specific point mutations in E. coli. gRNAs are delivered on a different plasmid and allows simultaneous multiplex editing of six different genes. This system doesn’t rely on any additional or host-dependent factors and can likely be used in a wide range of bacteria.
References
1. Maddalo, Danilo, et al. “In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system.” Nature 516(7531) (2014): 423-7. PubMed PMID: 25337876. PubMed Central PMCID: PMC4270925.
- Find plasmids from this paper at Addgene.
2. Kabadi, Ami M., et al. “Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector.” Nucleic Acids Research 42(19) (2014): e147. PubMed PMID: 25122746. PubMed Central PMCID: PMC4231726.
- Find plasmids from this paper at Addgene.
3. Sakuma, Tetsushi, et al. “Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system.” Scientific Reports 4 (2014): 5400. PubMed PMID: 24954249. PubMed Central PMCID: PMC4066266.
- Find plasmids from this paper at Addgene.
4. Albers, Joachim, et al. “A versatile modular vector system for rapid combinatorial mammalian genetics.” Journal of Clinical Investigation 125(4) (2015):1603-19. PubMed PMID: 25751063. PubMed Central PMCID: PMC4396471.
- Find plasmids from this paper at Addgene.
5. Tsai, Shengdar Q., et al. “Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing.” Nature Biotechnology 32(6) (2014): 569-576. PubMed PMID: 24770325. PubMed Central PMCID: PMC4090141.
- Find plasmids from this paper at Addgene.
6. Xing, Hui-Li, et al. “A CRISPR/Cas9 toolkit for multiplex genome editing in plants.” BMC Plant Biology 14 (2014): 327. PubMed PMID: 25432517. PubMed Central PMCID: PMC4262988.
- Find plasmids from this paper at Addgene.
7. Ma, Xingliang, et al. “A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants.” Molecular Plant 8(8) (2015): 1274-1284. PubMed PMID: 25917172.
- Find plasmids from this paper at Addgene.
8. Xie, Kabin, Minkenberg, Bastian, & Yinong Yang. “Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system.” Proceedings of the National Academy of Sciences USA 112(11) (2015): 3570-5. PubMed PMID: 25733849. PubMed Central PMCID: PMC4371917.
- Find plasmids from this paper at Addgene.
9. Yin, Linlin, et al. “Multiplex Conditional Mutagenesis Using Transgenic Expression of Cas9 and sgRNAs.” Genetics 200(2) (2015): 431-441. PubMed PMID: 25855067.
- Find plasmids from this paper at Addgene.
10. Port, Fillip, et al. “Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila.” Proceedings of the National Academy of Sciences USA 111(29) (2014): E2967-2976. PubMed PMID: 25002478. PubMed Central PMCID: PMC4115528.
- Find plasmids from this paper at Addgene.
11. Cress, Brady F., et al. “CRISPathBrick: Modular Combinatorial Assembly of Type II-A CRISPR Arrays for dCas9-Mediated Multiplex Transcriptional Repression in E. coli.” ACS Synthetic Biology 4(9) (2015):987-1000. PubMed PMID: 25822415.
- Find plasmids from this paper at Addgene.
12. Ronda, Carlotta, et al. (2016). "CRMAGE: CRISPR Optimized MAGE Recombineering." Sci Rep. 6:19452. PubMed PMID: 26797514
- Find plasmids from this paper at Addgene.
Additional Resources on the Addgene Blog
- Browse Our CRISPR Topic Page
- Get Advice on Choosing The Proper Cas9 Variant for Your Experiment
- Learn About Multiplexing with Cpf1
Additional Resources on Addgene.org
- Catch Up on Your CRISPR Background with Our Guide Pages
- Find CRISPR Plasmids for Your Research
- Browse Our CRISPR Multiplexing Resources
Topics: CRISPR, CRISPR 101, CRISPR gRNAs






Leave a Comment