Updated mini-transposon vector for bacterial mutagenesis or gene targeting
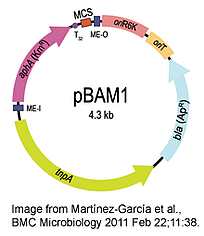
Victor de Lorenzo's lab has engineered a modular mini-Tn5 vector that can be used to generate random mutagenesis libraries or to insert heterologous genes, reporters, or other markers into a target genome. They did this by selecting the important elements from existing transposon and vector systems and creating an all-synthetic vector that included only the elements needed for function.
The lab validated this vector, called pBAM1, by conducting random mutagenesis in the soil bacterium Pseudomonas putida and demonstrate that they can successfully create GFP fusion proteins with a variety of genes across the genome. Although this tool was published in 2011, it was only recently made available through Addgene and we want to highlight it for use in your research.
-
Martínez-García et al., BMC Microbiology 2011 Feb 22;11:38.
Lentiviral CRISPR libraries for knockout screening
New systems have been developed and deposited with Addgene which allow scientists to use CRISPR-Cas technology to perform genome-wide knockout screens. These vectors and sgRNA libraries expand upon the CRISPR family of plasmids by offering a lentivirus-based mechanism for sgRNA delivery and providing a means for large scale functional screens.
For more information on these new CRISPR screening tools, see our CRISPR/Cas Plasmids: Pooled Libraries webpage or read our blog post,Lentiviral CRISPR Libraries Enable Genome-Scale, Knockout Screening.
Also, check out our updated CRISPR-Cas resources at www.addgene.org/CRISPR/ to browse CRISPR plasmids, watch informational videos, download protocols, and more! Looking for background information on CRISPR technology? See our improved CRISPR-Cas Guide.
-
Wang et al., Science. 2014. Jan 3;343(6166):80-4.
-
Shalem et al., Science 2014. Jan 3;343(6166):84-7.
-
Koike-Yusa et al., Nat Biotech. 2013. Dec 23.

Highly bright and extremely dim GFPs
The availability of the three dimensional structure of GFP, its brighter mutant S65T-GFP, and the subsequent biochemical characterization, has enabled scientists to engineer a wide variety of GFPs with diversity in fluorescence brightness, intensity, excitation and emission spectra. The lab of Dimitri Deheyn recently identified a family of GFP proteins in the cephalochordate Branchiostoma floridae, named bfloGFPs. They went on to characterize the structural and spectral properties of two of the family members – one with an unprecedentedly-high brightness, bfloGFPa1, and the other with extremely-dim brightness, bfloGFPc1. The plasmids used for determining the structures are now available from Addgene - bfloGFPa1 and bfloGFPc1.
- Bomati et al., Sci Rep 2014 Jun 27;4:5469.
MitoTimer plasmids to report mitochondrial turnover
Mitochondrial health, transport and turnover are important features to be monitored to study mitochondrial dysfunction. Easy-to-use tools to monitor these features were lacking until Roberta Gottlieb and Zhen Yan labs developed MitoTimer plasmids.
MitoTimer proteins are derived from the Timer protein - a mutant of the fluorescent protein dsRed that changes irreversibly its color from green to red when it is oxidized. The Timer tool has already been used by several groups to monitor protein and cell ageing, as the spectrum shift occurs during Timer protein life due to a form of oxidation (dehydrogenation). MitoTimers are composed of a Timer protein fused to the mitochondrial targeting sequence of cytochrome c oxidase subunit VIII, so that they can be imported to the mitochondrial matrix. Gottlieb’s lab has used an inducible MitoTimer reporter using a Tet-on system (pTRE-Tight-MitoTimer) to show its usefulness in cell culture to report changes in mitochondrial turnover and transport. pMitoTimer from Yan’s lab, which is constitutively expressed, reports on the balance of biogenesis and mitochondrial degradation in vitro and in vivo.
Merged green and red channel fluorescence images of C2C12 cells stably expressing pTRE-Tight-MitoTimer and rtTA. The cells were pulsed with 2µg/ml of Doxycycline for 2 hours, and imaged on the Keyence BZ-9000 fluorescence microscope 8 hours post-pulse. The distinct differences of red-to-green fluorescence in individual mitochondria in the networks marks the differences in import of newly synthesized (green) protein and organelles with older (yellow to orange) protein. Image courtesy of Roberta Gottlieb.
-
Laker et al., J Biol Chem 2014 Apr 25;289(17):12005-15.
-
Hernandez et al., Autophagy 2013 Nov 1;9(11):1852-61.
A quicker method for protein inactivation
Your protein of interest does x, but it also participates in y. Depletion of your protein may upregulate an alternative pathway or induce other compensatory mechanisms. How can protein function be dissected while minimizing confounding secondary effects?
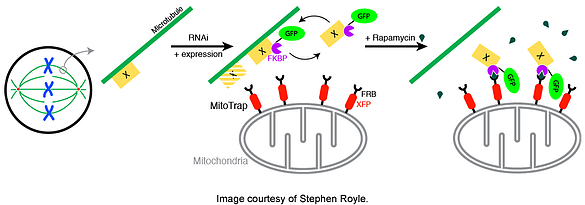
An approach described as “knocksideways” (a British idiom for “taking by surprise”) by Margaret Robinson’s lab, and more recently used by Stephen Royle’s lab, acutely depleted proteins from their compartment of action via sequestration.
The strategy relies on several observations: Rapamycin is a membrane-permeable pharmacological agent that can rapidly induction heterodimerization of proteins that contain rapamycin-binding domains, such as FKBP and FRB. The cell does not appear to mind extraneous proteins bound to its outer mitochondrial membrane.
Sequestration is accomplished by knocking down your endogenous protein of interest and co-expressing its replacement recombinant protein with a FKBP domain and a mitochondrial outer membrane protein with a FRB domain (MitoTRAP). Rapamycin binds your recombinant protein (via FKBP) and then sequesters the protein at the mitochondria (binding FRB), allowing for a rapid and inducible inactivation. Find the Robinson plasmids or the Royle plasmids and get to work!
-
Robinson et al., Dev Cell 2010 Feb 16;18(2):324-31.
-
Cheeseman et al., J Cell Sci 2013 May 1;126(Pt 9):2102-13.
Assemble multiple component plasmids from building blocks via Golden GATEway Cloning
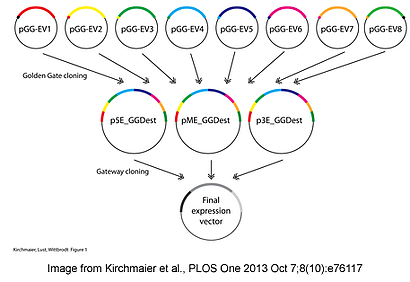
Frustrated by the complexity of assembling recombination of transgenesis constructs? A new cloning system combining the advantages of Golden Gate and Multisite Gateway cloning methods developed by the lab of Joachim Wittbrodt might be just the set of tools that you need.
The Golden GATEway cloning kit simplifies the cloning process for complex DNA constructs, particular for those involving recombination elements such as FRT or Lox sites, and provides a modular system for easier exchange and re-use of existing elements. DNA sequences for a desired component are cloned into Golden Gate entry vectors via traditional restriction enzyme cloning, TA cloning, or annealing of oligonucleotides. These entry vectors are used in a Golden Gate cloning step to assemble the individual components into a destination vector in a predefined order. Finally, the destination vectors are used with Multisite Gateway cloning to generate the final construct.
Up to eight entry vectors can be used for each Multisite Gateway compatible destination vector for a maximum of 24 elements assembled into a final transgenesis construct. Plasmids constructed using Golden GATEway cloning have been utilized to create recombination template vectors, to perform multiple site mutagenesis and create complex fusion or recombination vectors. The efficient and flexible cloning process provides improvements to classical cloning methods, particularly for complex transgenesis constructs.
-
Kirchmaier et al., PLOS One 2013 Oct 7;8(10):e76117.
Irreversible peptide-peptide ligation using SpyLigase
Building off their SpyTag/SpyCatcher system for protein-peptide locking, Mark Howarth’s lab has developed a new tool for peptide-peptide locking. The new technology is known as SpyLigase and is a protein domain that promotes the formation of an isopeptide bond between 2 peptide tags, SpyTag and KTag. The group demonstrated the use of the SpyLigase peptide-peptide interaction to link affibodies or antibodies against common tumor markers to enhance cancer cell capture.
For further reading about SpyLigase technology, read the Addgene interview with Mark Howarth.
Shining a light on channelrhodopsins – Chronos & Chrimson
The paper describes five previously unknown channelrhodopsins from different species: Stigleoclonium helveticum (ShChR), Chlamydomonas noctigama (CnChR1), Chloromonas subdivisa (CsChR), Chloromonas oogama (CoChR), and Scherffelia dubia (SdChR). ShChR, nicknamed Chronos, is blue and green light-excitable and has faster kinetics than those of other channelrhodopsins. CnChR1, nicknamed Chrimson, is the first reported yellow-peaked channelrhodopsin with a spectral peak at 590 nm, which is 45 nm more red-shifted compared to other variants. The group further optimized Chrimson through mutagenesis (K176R) to improve the otherwise slow off-kinetics (15.8 ± 0.4ms from 21.4 ± 1.1 ms); this new variant was named ChrimsonR. Plasmids containing Chronos, Chrimson, ChrimsonR, CsChR, and CoChR have been deposited to Addgene, including lentiviral and AAV expression vectors for certain variants.
-
Klapoetke et al., Nat Methods 2014 Mar 11;(3):338-46.
For more information and descriptions of various optogenetics plasmid tools, visit Addgene’s optogenetics guide.
More optogenetics tools – light-activated receptor tyrosine kinases
Receptor tyrosine kinases (RTKs) are a large class of cell-surface receptors which play a critical role in development and are often implicated in disease progression. One of the major challenges in signaling research is the inability to replicate the spatio-temporal precision with which signaling events occur in a physiological setting. Genetic techniques typically rely on overexpression or knock-down, where signaling dynamics are influenced by the relatively long life-cycle of a protein from expression to degradation. Chemical approaches may require expensive peptides or small molecules and exceed a physiologically relevant exposure in intensity and/or duration. Both of these strategies carry the risk of off-target effects.
Harald Janovjak and his team at the Institute of Science and Technology Austria, decided to take a novel approach to control for these effects, drawing on the rapidly growing field of optogenetics. Using a rational protein engineering approach, they designed Opto-RTKs, which activate signaling cascades on exposure to low-intensity blue light. This approach relies on the incorporation of light-oxygen-voltage (LOV) sensing domains from algae into chimeric Opto-RTKs. The LOV domains bind flavin cofactors and dimerize on exposure to light, bringing the intracellular kinase domains into contact and initiating signaling. Grusch et al. demonstrate that Opto-RTKs have a similar level of background activity and activation as their wild-type counterparts.
The Janovjak lab has deposited their 3 Opto-RTK constructs – Opto-mFGFR1, Opto-hEGFR, and Opto-hRET – as well as various LOV-domain-mVenus constructs used in the study for those who would like to extend this work into their RTK of interest.
-
Grusch, M. et al., EMBO J 2014 Aug 1;33(15):1713-26.
RUSH & CRUSH – rapid & conditional gene silencing in RNAi transgenic mice
-
Brown et al., Genesis 2014 Jan;52(1):39-48.
APEX2 for proteomic mapping and electron microscopy
A thorough understanding of complex biological systems requires both a catalog of molecular players and contextualized knowledge of their roles inside cells. The original APEX (enhanced ascorbate peroxidase) reporter from Alice Ting’s lab has enabled progress on both fronts – as a genetic electron microscopy (EM) tag providing superior subcellular localization of tagged proteins compared to traditional fluorescence microscopy, and as a targeted tool for spatial proteomic mapping in living cells. Despite these advances, the Ting lab’s first-generation APEX suffered from inconsistent activity levels and limited sensitivity under the stringently low expression conditions necessary to avoid biological perturbation in a variety of contexts. In order to overcome these limitations, they evolved and characterized an improved peroxidase reporter, APEX2, through yeast display. APEX2 has been shown to exhibit superior performance over APEX for both EM imaging and proximity-dependent proteomic mapping with far greater sensitivity at lower expression levels. APEX2 is a monomeric, 27 kD genetic tag which can be fused to proteins of interest for EM imaging, or targeted to subcellular regions or protein complexes for proteomic mapping in live cells.
-
Lam et al., Nat Methods 2014 Nov 24. doi: 10.1038/nmeth.3179.
-
Martell et al., Nat Biotechnol 2012 Nov;30(11):1143-8.
A new tool for CRISPR gRNA validation
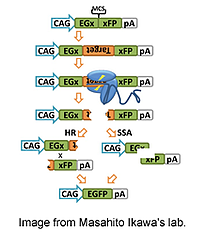
Selecting a gRNA sequence that effectively targets your gene/region of interest is a key step for CRISPR/Cas9 gene editing. Dr. Masahito Ikawa has created a GFP reporter plasmid for scientists to validate the efficacy of their gRNAs.
The first step is to clone the target sequence in between two fragments of EGFP in pCAG-EGxxFP. The EGFP fragments contain 482bp of overlapping sequence that direct Homologous Recombination (HR) or Single Strand Annealing (SSA) in the event of a DNA break. Next, the gRNA being tested is expressed along with Cas9. If the gRNA effectively cuts the target sequence, the plasmid undergoes HR or SSA to reconstitute functional GFP, and the cells will turn green.
-
Mashiko et al., Sci Rep 2013 Nov 27; 3:3355.
Rinehart lab reagents for improved expression of recombinant phosphoproteins
Protein phosphorylation is one of the most abundant forms of post-translational modifications in cells and research into its many roles in protein function and signaling networks continues to expand. The labs of Jesse Rinehart and Dieter Söll at Yale University previously changed the way researchers can explore important questions surrounding serine phosphorylation by adding this phosphorylated amino acid to the genetic code of E. coli (Park et al., Science 2011).
The Rinehart lab has now made improvements to this system by engineering cells that lack release factor one (RF-1; Bacterial strain EcAR7) and minimizing the set of plasmids required to make singly or multiply phosphorylated proteins (B40 OTS and pCRT7-GFP). To demonstrate the improvements of the system, the Rinehart lab synthesized the activated form of human mitogen-activated ERK activating kinase 1 (MEK1) with either one or two phosphoserine residues cotranslationally inserted in their canonical positions (SP218, SP222) using the original or improved phosphoprotein synthesis reagents. One phosphoserine (SP218) insertion was moderately enhanced while two phosphoserine insertions (SP218/SP222) was dramatically enhanced with the improved system. This MEK vector is also available at Addgene and can be used as a control for your experiments. For more information, please see Addgene’s information page for the Rinehart phosphoprotein system, which includes detailed protocols provided by the Rinehart lab.-
Steinfeld et al., ACS Chem Biol 2014 May 16;9(5):1104-12.
-
Heinemann et al., FEBS Lett 2012 Oct 19;586(20):3716-22.
-
Park et al., Science 2011 Aug 26;333(6046):1151-4.
pOSIP and the Clonetegration
Classic genetic engineering methods enabling chromosomal integration of sequences in bacteria are time-consuming and involve many steps. The Drew Endy and Keith Shearwin labs have developed a new, streamlined approach to genetic engineering which drastically reduces the time and effort needed to insert new genes into bacteria. They designed the pOSIP (one-step integration plasmid) series of plasmids, vectors that convey both the sequence to be integrated and a removable integrase cassette. They validated this methodology in two differents types of bacteria (E. coli and Salmonella typhimurium) by integrating DNA sequences either sequentially or simultaneously.
This method, called by the authors the “Clonetegration”, is quick and easy to do. Clonetegration could become a “valuable technique facilitating genetic engineering with difficult-to-clone sequences and rapid construction of synthetic biological systems” as they predict. The pOSIP plasmid kit can be found at Addgene, so what are you waiting for? Start building up your own designer bacteria.
-
St-Pierre et al., ACS Synth Biol 2013 Sep 20;2(9):537-41.
DREADD-based chemogenetic technologies
After several years of distributing his Designer Receptors Exclusively Activated by Designer Drugs (DREADD) plasmids on his own, UNC-Chapel Hill's Bryan Roth has now deposited many of these constructs with Addgene. These G-protein coupled receptors have been engineered by the Roth lab to be activated by otherwise pharmacologically inert drug-like small molecules, allowing labs to precisely and non-invasively control neuronal signaling.
For more information on DREADDs, please see the Roth lab's DREADD users blog.
New CRISPR plasmids available!
Interested in purifying Cas9? Check out pET-28b-Cas9-His from Alex Schier's lab, designed for expression and purification of Cas9 protein in Rosetta E. coli.New Lentiviral CRISPR activator and repressor plasmids from Scot Wolfe's lab. These include Tet-inducible CRISPR activators and repressor plasmids. (Kearns et al., Development. 2014..)
CRISPRs for Xenopus! From the lab of Yonglong Chen, pCS2-3xFLAG-NLS-SpCas9-NLS is a Cas9 expression plasmid that was used by Chen and colleagues for genome editing inXenopus tropicalis. (Guo et al., Development. 2014..)A new, higher specificity genome editing system that combines TALENs and CRISPRS. Developed by David Liu and colleagues, FokI-dCas9 expresses Fok1 nuclease domain fused to catalytically inactive Cas9 DNA-binding domain in mammalian cells. (Guilinger et al., Nat Biotechnol. 2014..)
Our CRISPR-Cas collection of plasmids updates frequently, so visit our CRISPR pages often to find the most recently deposited tools, resources, protocols, and more.
MoClo modular cloning system
Synthetic biologists have developed a modular cloning strategy, MoClo, which uses the Type IIS restriction enzymes BsaI and BpiI/BbsI to efficiently assemble up to six DNA fragments at a time. This method (based on the Golden Gate technology) exploits the ability of Type IIS enzymes to cut outside their recognition site, and permits DNA fragments with compatible overhangs to be efficiently assembled. Scientists can engineer unique enzyme recognition sites that flank a DNA module in an inverse orientation, so that multiple DNA components can directionally assemble in a single reaction, while retaining only a defined 4bp fusion site in between.
The MoClo system is comprised of three sets of cloning vectors (Level 0, 1, or 2) which can be utilized in three successive assembly steps. Before beginning, scientists can insert fragments of DNA containing basic parts (promoters, UTRs, coding sequences, terminators, etc) into individual Level 0 plasmids, or choose from a growing number of libraries containing pre-constructed standardized modules. In the first assembly step, compatible Level 0 vectors are directionally assembled into a Level 1 vector creating a single transcriptional unit (Ex: a promoter, 5’UTR, coding region, and terminator). Next, up to six Level 1 modules can be similarly assembled into a Level 2 vector, thus forming a functional genetic circuit. Flexibility has been built into the Level 2 vectors to allow for additional iterations of Level 1 assembly if necessary. Combining multiple Level 2 vectors in the final assembly step permits the creation of more complex constructs constrained only by the ability of E. coli to maintain the final plasmid after transformation.
Addgene depositors Sylvestre Marillonnet and Nicola Patron have assembled two collections of standardized genetic modules compatible with the MoClo system. The MoClo Toolkit provided by the Marillonnet Lab can be used to assembly general eukaryotic multigene constructs, while the Patron Lab MoClo Plant Parts kit contains modules specific for plant transformation.
-
Weber et al., PLoS One. 2011. Feb 18;6(2):e16765.
-
Engler et al., ACS Synth Biol 2014. Feb 5 (Web); DOI: 10.1021/sb4001504.
For further reading about Nicola Patron's MoClo kit and her plant synbio research, read the Addgene interview.
Engineering TALENs containing variable-repeats using Platinum Gate system
Interested in optimizing TALEN assembly and activity? The laboratory of Takashi Yamamoto has created a complete TALEN assembly system after systematically analyzing the effect of both the TALE scaffold and module on TALEN activity in a single-strand annealing (SSA) assay.
This new Platinum Gate TALEN Kit utilizes a 4-module assembly system, which reduces the number of individual repeat-variable di-residue (RVD) module plasmids and simultaneously increases the success rate of module assembly in the first Golden Gate reaction. While the DNA-binding specificity is imparted by the RVD at residues 12 and 13 in the TALE repeat, other naturally-occurring variations in the TALE repeat, referred to as “non-RVD variations”, were found to improve TALEN activity. Each positional group of module plasmids (ex. p1HD, p1NG, p1NI & p1NN) in the Platinum Gate system contains an identical variable repeat (VR) at the 4th and 32nd residues of the TALE repeat and the VR differs between the positional groups. Eight final destination vectors, consisting of all four final RVD modules in each of two different TALE scaffolds are provided, as the optimal scaffold can depend on the length of the spacer region between the target sequences. These updates to TALEN genome engineering demonstrate improved efficiency over previous reports.
For additional information on using the Platinum Gate TALEN Kit, please see the Yamamoto lab’s protocol for TALEN construction.
-
Sakuma et al., Sci Rep. 2013. Nov 29;3:3379.
Interested in more genome engineering technologies? Browse others on Addgene's Genome Engineering Guide.
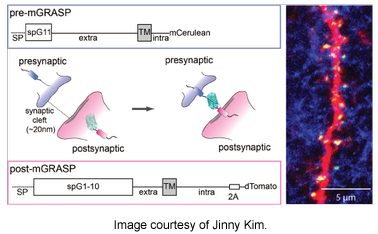
Fire up those neurons: mGRASP
A Nature 2012 article by Jinny Kim and colleagues describes their efforts to map the location and distribution of synapses in the mouse brain. Kim et al is utilizing a mammalian GRASP (GFP reconstitution across synaptic partners) technique based on functional complementation between two non-fluorescent split GFP fragments. When the two fragments, expressed in the presynaptic region of one neuron and postsynaptic region of a different neuron, come into proximity in the synaptic cleft, functional fluorescent GFP is reconstituted in vivo. Currently available from the Kim lab are 2 presynaptic and 2 postsynaptic targeting mGRASP plasmids. Additionally, the lab recently described the use of another set of mGRASP plasmids in their Neuron 2014 paper.
-
Kim et al., Nat Methods. 2011. Dec 4;9(1):96-102.
-
Druckmann et al., Neuron. 2014. Feb 5;81(3):629-40.
Next-Gen Brainbow toolkit for neuronal imaging
Joshua Sanes and his team at the Center for Brain Science at Harvard University have developed a next-generation Brainbow toolkit for high-resolution fluorescent imaging of individual neurons. The technology generates a unique spectral identity for each cell in a population by expressing a randomly generated mix of fluorescent proteins, determined by competing recombination events at the genetic level. Upon Cre/loxP recombination, each transgene expresses one of three possible fluorescent proteins, chosen for minimal spectral overlap, minimal protein aggregation, and high photostabilty. When multiple cassettes are integrated, each recombines independently, generating tens or hundreds of possible combinations (depending on the number of copies). This facilitates the distinction of neighboring cells in imaging applications and the mapping of neuronal projections to their associated cell bodies.
The first versions of the Brainbow system were reported in 2007. In order to extend the utility of the system, the researchers developed three new versions. Flipbow employs the Flp recombinase/FRT system in place of Brainbow’s Cre/loxP, allowing for simultaneous use of the two systems in different tissues of the same animal. Flipbow additionally incorporates SUMO tags in the FP sequence for separation from Cre-based Brainbow-expressing cells. Autobow plasmids are all-in-one versions which express self-excising Cre recombinase from the same transgene as the Brainbow cassette, simplifying experiments when additional cross-breeding steps are undesired or infeasible. Finally, an adeno-associated viral (AAV) system enables greater spatio-temporal control over expression and increases the number of species in which Brainbow may be used. This system uses two plasmids in tandem, with Cre-dependent inversion determining between 0 and 2 FPs expressed from each copy. Brainbow, Flipbow, and Autobow systems are available with either the Thy1 or CAG promoter, while Brainbow AAV is under control of the EF1a promoter.
-
Livet et al., Nature. 2007. Nov 1;450(7166):56-62.
-
Cai et al., Nat Methods. 2013. May 5;10(6):540-7.
pCoofy vectors for optimizing protein expression
Under the direction of Sabine Suppmann, the Recombinant Protein Production group at Max-Planck Institute of Biochemistry has developed a number of expression vectors for use with Sequence and Ligation Independent Cloning (SLIC). The pCoofy series of plasmids contain a variety of N- and C-terminal tags (including His, S-tag, OneStrep, CBP, Trx, GST, Halo, MBP, NusA and SUMO) for optimizing expression, solubilization and purification and have been tested in bacterial, insect and mammalian cells. These vectors were designed for parallel testing and screening of constructs in multiple host cells in order to optimize expression. The expression plasmids for a given species are based on the same backbone to permit expression levels to be directly compared among the different tags
To clone a sequence or gene of interest into the pCoofy vectors, select the appropriate pair of vector and gene primers from Table 2 of the associated publication which contain regions of sequence homology between the vector and gene primer for SLIC cloning. Amplify the pCoofy vector and the sequence of gene of interest in separate PCR amplication reactions for recombination in SLIC cloning to form the desired vector. The pCoofy vectors contain the ccdB cassette, which is not copied during the PCR amplification step, so that only vectors with the desired sequence of interest are retained after SLIC cloning. The general cloning strategy described in the associated publication can be used to generate any combination of tags for optimizing protein expression and purification in a fast, efficient and affordable way.
Scholz et al.., BMC Biotechnology. 2013. Feb 14;13:12.
GreenGate cloning system for plant transgenesis
Developed by Jan Lohmann and colleagues, GreenGate is a cloning system for the rapid assembly of plant transformation constructs. As the name suggests, GreenGate is based on the Golden Gate cloning method, but has been modified specifically to improve plant transgenesis. TheGreenGate kit available at Addgene includes six individual types of pre-cloned insert modules (plant promoter, N-terminal tag, coding sequence of the gene of interest, C-terminal tag, plant terminator, and plant resistance cassette) in pUC19 based entry vectors, as well as the pGreen-IIS based destination vectors.
To learn more about the GreenGate cloning system, see the detailed plasmid kit page or read our blog post: Quick, Versatile Plant Transgenesis with GreenGate Plasmids.
-
Lampropoulos et al., PLoS One. 2013. Dec 20;8(12):e83043.
What are your favorite plasmid technologies? Email us at blog@addgene.org and let us know what plasmids you would like to seen on our Hot Plasmid list.
Other popular plasmids & blog posts @Addgene:
Topics: Hot Plasmids, Other Plasmid Tools






Leave a Comment