Every few months we highlight a subset of the new plasmids and viral preps in the repository through our hot plasmids articles. These articles provide brief summaries of recent plasmid deposits and we hope they'll make it easier for you to find and use the plasmids you need. If you'd ever like to write about a recent plasmid deposit please sign up here.
 Here's what you'll find in this post:
Here's what you'll find in this post:
- Tools for labeling excitatory inhibitory synaptic proteins
- Fluorescent protein system for C. elegans
- Fluorescent timers for studying cell cycle length
- The CRISPR corner
- New from the viral service
Labeling excitatory and inhibitory synaptic proteins with FingRs
By Jason Nasse
Since the pioneering days of Golgi and Cajal, neuroscientists have developed and refined many tools to tease out the details underlying brain structure and function, with the advent of each tool expanding our ability to study and understand the complex interworking of the 86 billion neurons in the human brain. But studying synaptic connections and physiology is particularly challenging as they are great in number, generally very small, and can contain different neurotransmitters. Using genetically encoded fluorescent proteins coupled with Fibronectin intrabodies generated with mRNA display (FingRs), Xue Han’s lab has developed AAV vectors to selectively visualize excitatory and inhibitory synapses in the brain.
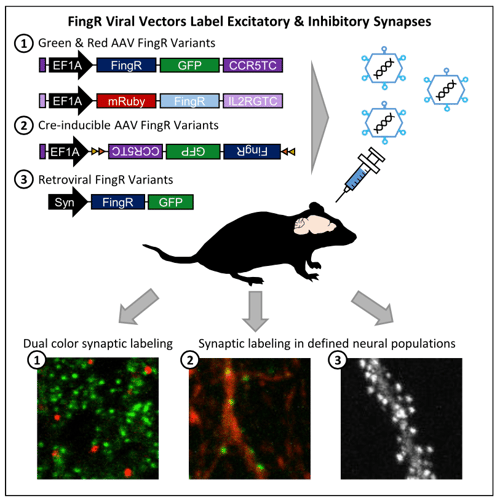 |
| Figure 1:Schematic diagram of different FingR constructs and experimental results. Image from Bensussen et al., 2020. |
The FingR constructs target the synaptic scaffolding proteins PSD95 and gephyrin to label excitatory and inhibitory synapses, respectively. Once expressed in the neuron, the FingRs bind to the endogenous synaptic proteins allowing them to localize to the synapse and minimize extraneous fluorescence from other regions of the cell. By combining vectors expressing different fluorescent proteins targeting inhibitory or excitatory synapses or that are Cre-inducible, researchers can explore synaptic integration and connectivity without the constraints of antibody labeling. This opens the door to in vivo experiments such as tracking the synaptic maturation pattern of hippocampal adult-born granule cells over time that were not previously possible.
Bensussen S, et al., iScience 2020. https://doi.org/10.1016/j.isci.2020.101330
Split-wrmScarlet, a fluorescent proteins system for Caenorhabditis elegans
Fluorescent proteins (FPs) traditionally introduced in Caenorhabditis elegans on extrachromosomal arrays can lead to artifacts due to overexpression. CRISPR can insert the FP gene into the target gene’s native locus, but introducing large genetic changes in C. elegans with CRISPR can be difficult since it requires the much less efficient double stranded templates, rather than the more efficient single-stranded oligodeoxynucleotide donors (ssODN). So how can you use CRISPR to introduce large FP genes into C. elegans? By using a split FP approach.
Split-wrmScarlet is a new split red FP that is three times brighter in worms than split-sfCherry3. It was created by Maria Ingaramo, Jerome Goudeau, Cynthia Kenyon, and colleagues at Calico Life Sciences. Like many split FPs, split-wrmScarlet is composed of two FP fragments that are not fluorescent on their own. When the two fragments come together, they fuse and fluoresce. The team engineered C. elegans cell lines to express wrmScarlet1-10 unattached to any other cellular protein. They used CRISPR with a ssODN template to introduce the small wrmScarlet11 which is less than 60 nt in length to tag the target protein. The team validated this tool by targeting six different genes whose protein products have distinct localizations.
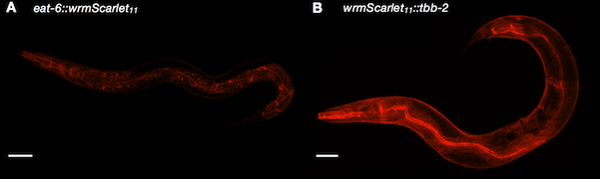 |
| Figure 2: Split-wrmScarlet labeling proteins with different subcellular locations. Image from Goudeau et al., 2020. |
Goudeau, J. bioRxiv 2020. https://doi.org/10.1101/2020.07.02.185249
Predicting cell cycle length with a histone-fused fluorescent timer
Studies of the cell cycle attempt to understand development, regeneration, and certain disease states. However, it’s difficult to observe the cell cycle in vivo, particularly if the cells are migratory or in deep tissues. To resolve this issue, Shangqin Guo and her colleagues at Yale University fused a color-changing fluorescent timer (FT) protein to the histone H2B that allows for observation in live cells. The fluorescent timer emits blue light around the time of synthesis and, after a few hours, a conformational change causes the protein to fluoresce red. H2B-FT-red is long lasting and was shown to have a half life of 84 hours in primary mouse embryonic fibroblasts. This fusion protein can be used to determine cell cycle length by examining the ratio of blue to red cells. Cells undergoing rapid cell cycles have a high blue to red ratio, whereas cells with long or slow cycles are more red than blue (Eastman et al., 2020).
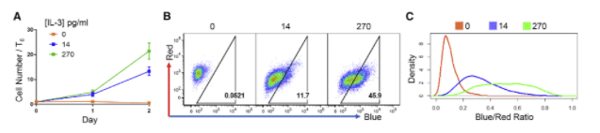 |
| Figure 3: BaF3 cells were transduced with H2B-FT virus and grown in varying concentrations of IL-3. Cells grown in high concentrations of IL-3 show a high proportion of blue fluorescence, indicating cell proliferation. BaF3 cells maintained in media without IL-3 show only red fluorescence. Image from Eastman et al., 2020. |
The researchers tested several scenarios from embryo development to myeloid proliferation in acute myeloid leukemia and saw that expression of H2B-FT consistently predicted the cell cycle speed across these different cell types. In one example, H2B-FT was expressed in murine B cells (BaF3), which depend on IL-3 to proliferate. The cells cultured with a low dose of IL-3 showed reduced proliferation and had a greater number of red cells as measured by flow cytometry. Cells grown in high IL-3 showed mostly blue fluorescence, indicating a quick cell cycle. Cells that had IL-3 withdrawn became entirely red as they could not proliferate.
Eastman et al. Cell Reports 2020. https://doi.org/10.1016/j.celrep.2020.107804
The CRISPR corner
Here are a few highlights from recent CRISPR plasmids. To find all of the CRISPR plasmids available from Addgene, head over to our CRISPR Plasmids and Resources page.
- CasPhi is 70 kDa Cas protein identified exclusively in huge bacteriophage genomes. It’s active in vitro and in human and plant cells. Its small size offers advantages for cellular delivery.
- Sc++ is a codon-optimized, engineered Streptococcus canis Cas9 nuclease that has a broad 5′-NNG-3′ PAM.
- The Human GlycoGene CRISPR Library is a new lentiviral CRISPR library that targets human glycosyltransferase genes and other proteins related to cellular glycosylation.
- CasRx is a RNA targeting Cas system that can be used in Drosophila melanogaster.
- The Cas Hybrid for Multiplexed Editing and screening Applications (CHyMErA) Knockout Libraries express hybrid Cas9-Cas12a gRNAs under a single U6 promoter. The paralog and dual-targeting hybrid gRNA library can be used to identify genetic interactions between paralogs. The exon-deletion hybrid gRNA library deletes human alternative cassette exons and disrupts genes with alternative exons.
New from the viral service
By Jason Nasse
We regularly add new viral aliquots from our plasmid collection to provide ready-to-use viral preps. Here are some of the new viral preps from recent months:
- The Dolcetto human CRISPR inhibition pooled library is now available as lentiviral preps in addition to the pooled library format! The library inhibits over 18,000 human genes.
- hSyn1-SIO-stGtACR1-FusionRed is a Cre-dependent, soma-targeting inhibitory channelrhodopsin. It’s now available as AAV5.
- The red-shifted calcium sensor jRGECO1a is now available as AAV-PHPeb.
Topics: Hot Plasmids, Other Plasmid Tools, Plasmids









Leave a Comment