Every few months we highlight a subset of the new plasmids in the repository through our hot plasmids articles. These articles provide brief summaries of recent plasmid deposits and we hope they'll make it easier for you to find and use the plasmids you need. If you'd ever like to write about a recent plasmid deposit please sign up here.
Human whole genome sgRNA iBAR library
Article contributed by Susanna Bachle
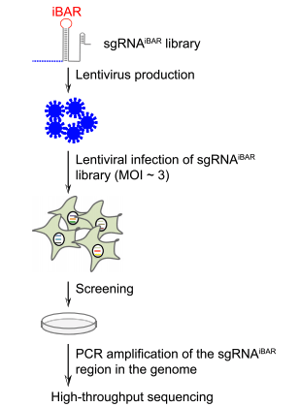 |
| Figure 1: Initial steps in the CRISPR-pooled screening using sgRNAiBAR. Image from Zhu et al., 2019. |
Wensheng Wei's lab at Peking University has developed a new CRISPR screening method that makes use of guide RNAs with internal barcodes (iBARs) in their loop region. After infecting cells with sgRNAiBAR library containing lentiviral vectors the iBARs can be detected by next generation sequencing.
In order to avoid a high false-positive rate discovery rate, conventional CRISPR screens require a low multiplicity of infection (MOI) during cell library construction to ensure that the majority of cells receive one gRNA. The iBAR method resulted in high quality data even when used at a high multiplicity of infection, because this setup allows for experimental replicates for each of the gRNAs. The idea was to assign four different barcodes to each of the gRNAs and to embed these barcodes within the gRNA sequences. This allowed tracing of the performance of each gRNA multiple times within the same experiment by counting both the gRNAs and their iBAR sequences. Since the sgRNAiBAR library covers every annotated human gene, it enables genome-wide screening with a high MOI, which is beneficial for a low number of cells and in vivo settings.
Zhu et al., Genome Biology, 2019. PubMed PMID: 30678704.
Sparse cell labeling using recombinase competition
Article contributed by Aliyah Weinstein
Sparse Predictive Activity through Recombinase Competition (SPARC) is a fully genetic tool for labeling just a fraction of a genetically-defined subset of cells. By labeling fewer cells in a population, it’s easier to visualize individual/non-overlapping cells. Until now, tools for doing this were labor-intensive and required using manipulations such as injection of precise titrations of AAV or heat-shocking the organism.
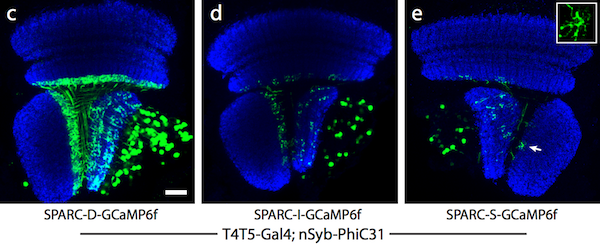
The SPARC system uses competitive recombination sites to selectively remove a stop cassette upstream of an effector/reporter gene. In cells where the stop cassette is removed, the effector is expressed. By truncating one of the recombination sites in the construct to different lengths, the density of cells that are labeled can be altered to dense (45% of cells labeled), intermediate (12%), and sparse (4%) levels.
SPARC is a versatile system that can be used in many cell types and organisms including Drosophila, fish, and mice, by replacing the effector with a gene of interest in the cells you are studying. Read our SPARC blog post for more details.
 |
|
Figure 2: Dense (c), intermediate (d), and sparse (e) labeling of the calcium indicator, GCaMP6f, in the Drosophila optic lobe. Image from Isaacman-Beck et al., 2019. |
Get started with sparse labeling
Isaacman-Beck et al., bioRxiv 2019. https://doi.org/10.1101/788679
A destabilized GFP reporter for calcineurin activity in yeast
Article contributed by Leah Schwiesow
Calcineurin is a protein phosphatase that responds to rising intracellular calcium levels within the cell. When levels of Ca2+ are high in the cell, calcineurin dephosphorylates the protein CZI, which in turn binds to calcineurin-dependent response elements (CDREs) and drives gene expression. Because calcineurin is required for virulence in pathogenic fungi, it is an attractive drug target. Calcineurin reporters allow scientists to measure calcineurin dynamics and can serve as a drug-screening platform to identify new antifungals that target calcineurin.
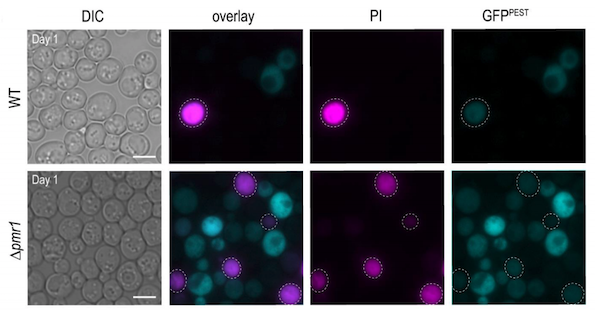
Current calcineurin gene reporters are based on the bacterial βgal assay, and are not suited to high throughput screening, calcineurin dynamics measurements, or monitoring in vivo. To overcome these technical issues, Sabrina Büttner’s lab designed a calcineurin transcriptional reporter consisting of a quickly-degraded destabilized GFP variant fused to a 4X CDRE repeat. In addition to showing that this vector is suitable for flow cytometry and fluorescence microscopy, they also show that this method is compatible with dead cell staining using propidium iodine (PI), allowing for calcineurin activity measurements in stressed or aging cell populations.
|
|
| Figure 3: Micrographs of wild type (top panels) and cells with high intracellular Ca2+ (bottom panels) equipped with destabilized GFP calcineurin reporter (blue) and stained with PI (pink) to indicate cell death. Image from Diessl et al., 2020. |
Find the GFP reporter calcineurin activity here
Diessl et al., Microbial Cell, 2020. https://dx.doi.org/10.15698%2Fmic2020.04.713
Truncated TALE-fluorescent protein fusions as a DNA staining dye
Article contributed by Jennifer Tsang
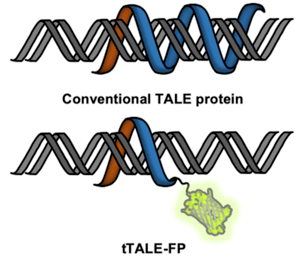 |
| Figure 4: Double stranded DNA bound by tTALE-FP with the fluorescent protein fluorescing green. Image from Shin et al., 2019. |
Looking for an alternative to intercalating fluorescent dyes for DNA-protein interaction studies? Kyubong Jo's lab generated fluorescent protein fusions with tTALE to “stain” DNA. TALE proteins can bind specific DNA sequences which brings the attached fluorescent protein to the DNA. They created plasmids for purification of tTALE-eGFP and tTALE-mCherry.
In comparison to intercalating fluorescent dyes which can cause photo-induced DNA breakage and structural deformation, these probes are reversible and do not cause structural deformation or photo-induced damage. These fusions can also function in high salt conditions - up to 100 mM NaCl. These tTALE-FPs also have higher DNA binding affinity for AT-rich regions vs. GC-rich regions from 40 mM to 100 mM NaCl but stained evenly in 1x TE buffer. The lab also used tTALE-FP in a single molecule-fluorescence assay to monitor restriction enzyme digestion of single molecules in real-time.
Find plasmids expressing tTALE-FP
Shin et al., Sci. Rep. 2019. https://doi.org/10.1038/s41598-019-53722-0
Topics: Hot Plasmids, Other Plasmid Tools, Plasmids






Leave a Comment