Adeno-associated virus (AAV) has many features which make it a great viral vector, but its packaging capacity is limited to ~4.7kb, or roughly half the packaging limits of lentiviral and adenoviral vectors. While many transgene will fit within this limit, some like prime editing's PE2 enzyme do not. So how do you fit a big gene into a tiny vector like AAV? By breaking the transgene into smaller pieces.
Reassembly of fragmented genomes is the key to fitting large genes into AAV
First, some history. In 2008, the Auricchio lab decided to solve the issue of “tiny vector, big transgene” by just straight up packaging an oversized genome of ~9 kb into AAV. They found that in defiance of the packaging limit dogma, this AAV successfully mediated transduction and expression of the full-length transgene in vitro and in vivo (Allocca et al., 2008). Shortly after though, three groups independently tried reproducing these results and determined that even when larger vector genomes are packaged, the physical size of the genomes is still ~4.7kb. But despite this, larger functional transgene products were produced following transduction.
So what’s going on here? The working model is that the oversized AAV genomes are fragmented or truncated when packaged and then following transduction, the partial genomes complement each other to restore the full-expression cassette. AAV’s natural ability to reassemble fragmented genomes is the basis of several split vector approaches developed in the early 2000’s which allow researchers to deliver oversized cargo (Hirsch et al., 2010).
Today’s blog post will give brief overviews of three split vector strategies plus one additional split vector strategy for increasing the size of the AAV donor template for CRISPR-mediated homology directed repairs (HDR).
Split AAV vector approaches
Split vector approaches increase the size of the gene delivered by splitting the gene into two pieces: part A and part B. Each piece of the gene is then independently packaged into an AAV. When a cell is co-infected with both part A and part B AAVs, the fragments reassemble and produce the full-length gene. Below are overviews and drawbacks of four split AAV vector approaches.
1. Overlapping
Overview: The overlapping strategy has a region of homology, typically ~400-1400 bp long between part A and B of the transgene to reconstitute the full-length transgene (Chamberlain et al., 2016). It’s generally accepted that some form of homologous recombination (HR) occurs via this region of overlap, but the exact mechanism is not yet known.
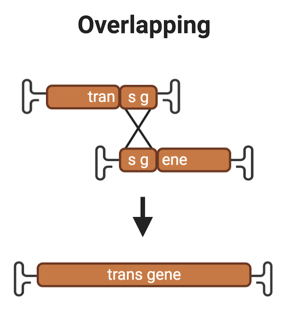
Drawbacks:
- Generally an overlapping split vector has equal or higher transgene expression when compared to using a single over-sized vector, but not always.
- The combination of serotype and tissue/cell type could have a big impact on the rate of transgene expression with an overlapping AAV since both of these variables influence whether cells will be co-transduced with multiple copies of both vectors.
Examples of use:
- Halbert et al., 2002 used an overlapping vector with 440 or 1000bp overlaps to deliver the alkaline phosphatase gene to airway epithelial cells in mice. This vector transduced ~10% of cells, similar to the transduction rate of a single AAV.
- Odom et al., 2011 used an overlapping vector with 372 bp overlaps to deliver the mini-dystrophin gene to a mouse model of muscular dystrophy. While mini-dystrophin expression levels were lower than expression of wild-type dystrophin in normal muscles, mice treated with the overlapping split vector had an improvement in muscle physiological performance.
2. Trans-splicing
Overview: For trans-plicing, splice site donor and acceptor sequences are used to reconstitute the two pieces of the transgene.The AAV encoding part A of the transgene contains a promoter and the 5’-fragment of the transgene, followed by a splice site donor. The AAV encoding part B of the transgene contains a splice site acceptor followed by the 3’-end of the transgene. When a cell is transduced by both viruses, if the AAV genomes encoding part A and part B of the transgene form a heterodimer, then splicing of the pre-mRNA of the two halves of the transgene will reconstitute the full-length transgene.
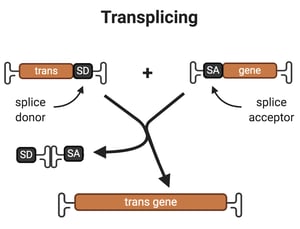
Drawbacks:
- Trans-splicing occurs across the ITR junction, and this is likely a rate limiting step, but optimization of the exon-splitting site sequence can enhance the rate of trans-splicing (Lai et al., 2005).
- Trans-splicing does not always outperform single oversized AAVs (Hirsch et al, 2013).
Examples of use:
- Lai et al., 2005 used a trans-splicing vector to deliver a 6 kb mini-dystrophin gene to a mouse model of muscular dystrophy. Three different splice site donor and acceptor sequences were tested and with the optimal sequences, the group achieved ~80% of muscle cells expressing mini-dystrophin.
3. Hybrid
Overview: The hybrid approach combines the overlapping and trans-splicing methods. The AAV encoding part A of the transgene contains the promoter, the 5’ half of the transgene, a splice donor sequence and the region of homology. The AAV encoding part B of the transgene contains the region of homology, a splice acceptor sequence, the 3’ half of the transgene. The full length gene is reconstituted by either the overlapping or the trans-splicing mechanism.
The splice site donor and acceptor sequences and the homologous region of the two vectors can be from one of the transgene’s endogenous introns or comprised of non-endogenous sequences, such as a 77bp long highly recombinogenic homology sequence derived from the filamentous phage F1 (Trapani et al., 2014).
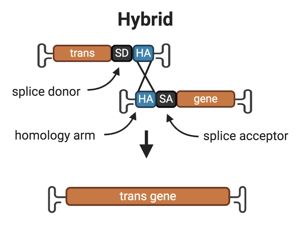
Drawbacks:
- The choice of homology sequence can influence the level of transgene expression, and may need to be optimized for each unique application (Trapani et al., 2015).
- Expression of smaller fragments of the transgene have been seen with the hybrid method. While these fragments are likely non-functional, they could be dominant-negative and/or toxic. A potential solution is to include an in-frame degron, or degradation sequence, upstream of the 5’ splice donor and downstream of the 3’ splice acceptor of the transgene to help prevent the accumulation of such truncated proteins.
Examples of use:
- Ghosh et al., 2008 used a hybrid split vector with a 872 bp long recombinogenic homology sequence to deliver LacZ to muscle in mice. Expression from the hybrid vector, as measured by β-galactosidase activity, was 81% of that from a single vector, but significantly greater than the expression from an overlapping (34%) or trans-splicing (62%) vector.
4. Sequential homology directed repairs
Overview: The sequential homologous recombination approach delivers large donor templates using two sequential homology directed repairs (HDR) with CRISPR/Cas9. Donor A contains: 400bp homology arms to the site in the genome being targeted for integration, “part A” of the transgene, the gRNA target site that mediated the donor template’s integration so that the site is reconstituted after integration; and a sequence of stuffer DNA after the gRNA target site.
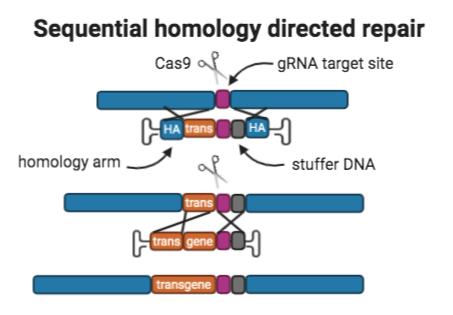
Donor B contains: 400 bp homology arms to donor A,and “part B” of the transgene.
These two AAVs are co-transduced in a single step and serve as donors for two consecutive CRISPR-mediated HDR events which results in the integration of a larger donor template.
Drawbacks:
- Transgene expression varies with this approach, ranging from ~10% expression in primary human T cells and hematopoietic stem and progenitor cells (HSPC)s to ~40% expression in the K562 cell line (Bak et al., 2017).
- Since this approach is dependent on double-strand DNA break repair, if non-homologous end joining (NHEJ) disrupts the gRNA target site after the first HDR event, then the second HDR event can’t occur.
- Lastly, depending on its design, donor A may be capable of expressing a truncated protein. This can be avoided by excluding a stop codon from donor A in any of the reading frames downstream of the gRNA target site so that transcripts undergo nonstop decay (Frischmeyer et al., 2002).
How to pick a split AAV method
While the hybrid dual vector system has a slight advantage over the overlapping and trans-splicing approaches since there are two ways for a transgene to be reconstituted, there is currently no one ideal split vector system. Overall the efficiency of transgene expression from split vectors has been inconsistent. A greater understanding of basic AAV biology, however, would help improve split vector strategies and lead to greater and more consistent transgene expression.
References:
Allocca M, Doria M, Petrillo M, Colella P, Garcia-Hoyos M, Gibbs D, Kim SR, Maguire A, Rex TS, Di Vicino U, Cutillo L, Sparrow JR, Williams DS, Bennett J, Auricchio A (2008) Serotype-dependent packaging of large genes in adeno-associated viral vectors results in effective gene delivery in mice. J Clin Invest 118:1955–1964 . https://doi.org/10.1172/jci34316
Bak RO, Porteus MH (2017) CRISPR-Mediated Integration of Large Gene Cassettes Using AAV Donor Vectors. Cell Reports 20:750–756 . https://doi.org/10.1016/j.celrep.2017.06.064
Carvalho LS, Turunen HT, Wassmer SJ, Luna-Velez MV, Xiao R, Bennett J, Vandenberghe LH (2017) Evaluating Efficiencies of Dual AAV Approaches for Retinal Targeting. Front Neurosci 11: . https://doi.org/10.3389/fnins.2017.00503
Chamberlain K, Riyad JM, Weber T (2016) Expressing Transgenes That Exceed the Packaging Capacity of Adeno-Associated Virus Capsids. Human Gene Therapy Methods 27:1–12 . https://doi.org/10.1089/hgtb.2015.140
Frischmeyer PA (2002) An mRNA Surveillance Mechanism That Eliminates Transcripts Lacking Termination Codons. Science 295:2258–2261 . https://doi.org/10.1126/science.1067338
Ghosh A, Yue Y, Lai Y, Duan D (2008) A Hybrid Vector System Expands Adeno-associated Viral Vector Packaging Capacity in a Transgene-independent Manner. Molecular Therapy 16:124–130 . https://doi.org/10.1038/sj.mt.6300322
Halbert CL, Allen JM, Miller AD (2002) Efficient mouse airway transduction following recombination between AAV vectors carrying parts of a larger gene. Nat Biotechnol 20:697–701 . https://doi.org/10.1038/nbt0702-697
Hirsch ML, Agbandje-McKenna M, Jude Samulski R (2010) Little Vector, Big Gene Transduction: Fragmented Genome Reassembly of Adeno-associated Virus. Molecular Therapy 18:6–8 . https://doi.org/10.1038/mt.2009.280
Hirsch ML, Li C, Bellon I, Yin C, Chavala S, Pryadkina M, Richard I, Samulski RJ (2013) Oversized AAV Transductifon Is Mediated via a DNA-PKcs-independent, Rad51C-dependent Repair Pathway. Molecular Therapy 21:2205–2216 . https://doi.org/10.1038/mt.2013.184
Lai Y, Yue Y, Liu M, Ghosh A, Engelhardt JF, Chamberlain JS, Duan D (2005) Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nat Biotechnol 23:1435–1439 . https://doi.org/10.1038/nbt1153
Odom GL, Gregorevic P, Allen JM, Chamberlain JS (2011) Gene Therapy of mdx Mice With Large Truncated Dystrophins Generated by Recombination Using rAAV6. Molecular Therapy 19:36–45 . https://doi.org/10.1038/mt.2010.205
Trapani I, Colella P, Sommella A, Iodice C, Cesi G, Simone S, Marrocco E, Rossi S, Giunti M, Palfi A, Farrar GJ, Polishchuk R, Auricchio A (2013) Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol Med 6:194–211 . https://doi.org/10.1002/emmm.201302948
Trapani I, Toriello E, de Simone S, Colella P, Iodice C, Polishchuk EV, Sommella A, Colecchi L, Rossi S, Simonelli F, Giunti M, Bacci ML, Polishchuk RS, Auricchio A (2015) Improved dual AAV vectors with reduced expression of truncated proteins are safe and effective in the retina of a mouse model of Stargardt disease. Hum Mol Genet 24:6811–6825 . https://doi.org/10.1093/hmg/ddv386
Additional resources on the Addgene blog:
- Find tips for a first time AAV user
- Learn about how AAV is used in retinal gene therapy
- Learn about the parts of an AAV transfer plasmid
Resources on Addgene.org:
Topics: Viral Vectors, AAV







Leave a Comment