When you think of antibodies, you probably think of monoclonal antibodies (mAbs). mAbs have been around since 1975 thanks to the Nobel prize winning work of Milstein and Köhler who developed hybridoma technology. But what about single chain fragment variables (scFvs)? scFvs weren’t developed for another decade when in 1988 they were cloned by two separate labs (Bird et al., 1988; Huston et al., 1988). scFvs are the smallest unit of an antibody molecule that can bind antigen. While they may seem unfamiliar, scFvs are often the building blocks for engineering proteins. So let’s get acquainted with scFvs.
What is an scFv?
scFVs are a type of recombinant antibody. They are ~25 kDa single polypeptides that contain the variable light chain (VL) and variable heavy chain (VH) of an antibody. These two chains are connected by a flexible linker peptide that is usually 15-20 amino acids long and made up of glycine and serine with dispersed hydrophilic residues for increased solubility (Monnier et al., 2013). The linker keeps the C-terminus of one variable domain and the N-terminus of the other domain at a distance that favors proper folding and formation of the antigen-binding site while also minimizing oligomerization of the scFv. While you might assume that the variable domains of an scFv would mirror that of an antibody (VL-linker-VH), both VL-linker-VH and VH-linker-VL configurations can generate functional scFvs; however, some individual scFvs performing better in one configuration than the other (Sandomenico et al., 2020).
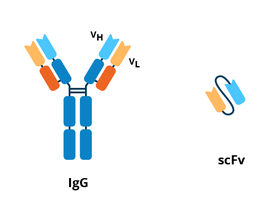 |
| Figure 1: Comparison between the IgG antibody and scFv. |
What are the advantages and disadvantages of a scFv vs. a monoclonal antibody?
Advantages of scFvs:
scFv’s advantages are rooted in their size. Being smaller means scFvs are easier and cheaper to make because they can be expressed in bacteria, while mAbs generally require mammalian expression systems. scFvs are also small enough to be screened for with in vitro display methods such as phage display (Bradbury et al., 2011). In vitro selection avoids animal immunization and also allows for generations of scFvs against two antigens that are impossible to generate scFvs against in vivo: antigens found in the body or self antigens, and toxins that are lethal to animals.
In the clinic, scFvs’ size also provides advantages over antibodies (Ahmed et al., 2012, Bates and Power., 2019): rapid blood clearance, which is useful for imaging applications; better tissue penetration which is useful for therapeutic and imaging applications; and reduced immunogenicity when administered in vivo due to their lack of an Fc region.
Disadvantages scFVs:
Compared to antibodies, scFvs tend to have lower affinities, lower long-term stability, and a higher likelihood to aggregate due to their small size (Bates and Power., 2019). Their rapid clearance from blood can be a drawback for therapeutic applications where longer retention times often increase therapeutic efficacy (Ahmed et al., 2012).
How do you generate an scFv?
scFv sequences have traditionally been cloned by amplifying the VLand VH antibody sequences of hybridomas, but phage display is a popular way to generate scFvs.
For phage display, a pooled library of scFvs is displayed on the coats of bacteriophages, a process which links genotype and phenotype. Incubating the phage display library with plate-bound antigen helps select for high affinity scFvs. Bound phage are then proliferated by infecting bacteria for additional rounds of panning and scFv characterization.
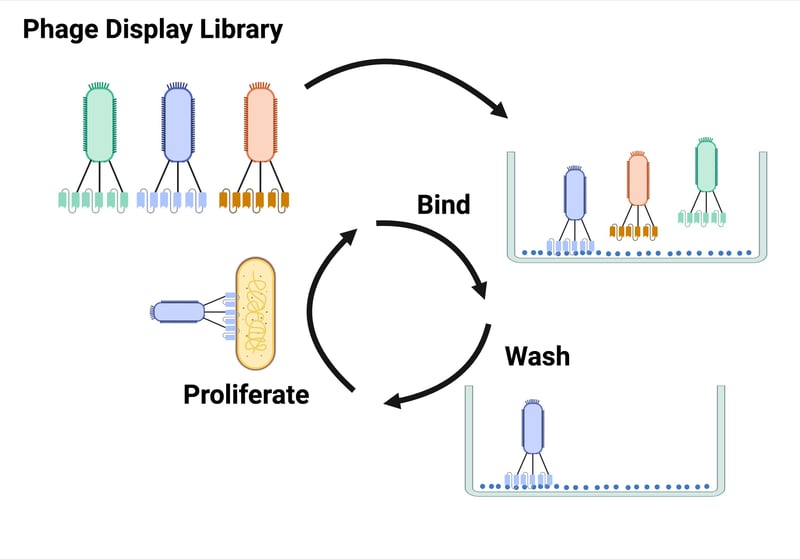 |
| Figure 2: Phage display starts with a pooled library of scFvs that are displayed on the coats of bacteriophages. This library is incubating with plate-bound antigen to select for scFvs with high affinity binding. Bound phage are then proliferated by infecting bacteria for additional rounds of screening. Created with Biorender.com. |
Some phage display libraries screen antibody genes from B cells of immunized animals, but it's possible to avoid animal use by using semi-synthetic or entirely synthetic VL and VH sequences. Phage display also allows for screening scFv generated from the B cell antibody genes from diseased or vaccinated humans.
How do you produce scFvs?
While it’s possible to produce scFvs in mammalian, yeast, plant, and insect cells, they are most often expressed from a plasmid in bacteria (Ahmed et al., 2012). One challenge of bacterial expression is the proper formation of the disulfide bond between the VL and VH domains. This bond provides stability and solubility for the scFv (Gaciarz and Ruddock., 2017), but requires an oxidizing environment to form. When expressed in the non-oxidizing cytoplasm of bacteria, scFvs accumulate in insoluble inclusion bodies. While it’s possible to re-solubilize and re-fold scFvs from inclusion bodies, it’s time-consuming and laborious.
To avoid this, one or more of these approaches are often used for scFv expression in bacteria:
- targeting to the oxidizing periplasm of bacteria to allow for proper disulfide bond formation.
- expression in strains of bacteria that are redox mutants so they have a more oxidizing cytoplasm.
- expression in the presence of molecular chaperones that help scFvs fold properly in the cytoplasm (Sandomenico et al., 2020).
- conversion to a cysteine free format that’s easier to express in the cytoplasm of E. coli, although this is not possible for all scFvs.
What are scFvs used for?
scFvs are used in many of the ways antibodies are. scFvs are even used to screen for new antibodies. To do this, antibody variable domain sequences are expressed as scFvs and screened with in vitro assays to select for strong binders. Then the sequences for the variable domains of the selected scFvs are inserted into a vector encoding the antibody constant domain scaffold to convert the scFv into an antibody.
The ability to combine scFvs with other protein domains, much like lego pieces, makes them better suited than antibodies to be used as building blocks for engineered proteins. This flexibility is largely due to scFvs being smaller than antibodies. scFvs are often genetically fused to other proteins, which has led to their application for basic and translational research.
Basic research applications of scFvs:
- The SunTag system uses an scFv to amplify the fluorescence intensity of a tagged protein. SunTag has two components: 1) a protein of interest that’s tagged with 10-24 copies of the short epitope GCN4, also called a scaffold; and 2) a GCN4 binding scFv that’s fused to GFP. When the multiple copies of the scFv-GFP fusion bind the SunTag scaffold, the intensity of the fluorescent signal is boosted and enables single molecule tracking in living cells.
- The HA frankenbody is an HA tag detection probe. It contains an HA-binding scFv that’s fused to a fluorescent protein such as GFP. The HA frankenbody works just like an antibody-based probe, but is easier and cheaper to generate since it’s genetically encoded.
Translational applications of scFvs:
- Just like antibodies, there are many scFv-based therapies in clinical trials or already in the clinic (Bates and Power., 2019).Many of the scFvs designed for oncology applications are reformatted as bispecific molecules which bind CD3 and a tumour specific antigen. When these molecules, called Bispecific T-cell engagers (BiTE®s), bind CD3 on T cells and a tumor-specific antigen, it brings T cells to a tumor site. ScFvs can also be fused to cellular toxins, radioisotopes, cytokines, and enzymes for cancer, autoimmune, and/or inflammatory therapeutic applications. Antibody fragments such as scFvs are also used for eye diseases, such as age-related macular degeneration (AMD) since direction injection to the eye results in high drug concentrations in the eye with minimal systemic side effects.
- scFvs are part of engineered chimeric antigen receptors (CARs). CARs are a chimera of scFvs and T cell receptors and are expressed on T cells to make CAR T cells. The extracellular scFv portion of the receptors recognizes an antigen of interest while the intracellular T cell receptor portion of the CAR facilitates signal transduction and release of cytotoxic granules from the T cell. When a CAR has an scFv that binds cancer-related antigens, binding of the scFv part of the receptor can activate T cells to kill cancer cells.
- scFv’s small size also allows for their delivery by viral vectors like AAV, which could be useful for delivering scFvs that inhibit HIV.
References and Resources
References:
Ahmad ZA, Yeap SK, Ali AM, Ho WY, Alitheen NBM, Hamid M (2012) scFv Antibody: Principles and Clinical Application. Clinical and Developmental Immunology 2012:1–15 . https://doi.org/10.1155/2012/980250
Bates A, Power CA (2019) David vs. Goliath: The Structure, Function, and Clinical Prospects of Antibody Fragments. Antibodies 8:28 . https://doi.org/10.3390/antib8020028
Bird R, Hardman K, Jacobson J, Johnson S, Kaufman B, Lee S, Lee T, Pope S, Riordan G, Whitlow M (1988) Single-chain antigen-binding proteins. Science 242:423–426 . https://doi.org/10.1126/science.3140379
Bradbury ARM, Sidhu S, Dübel S, McCafferty J (2011) Beyond natural antibodies: the power of in vitro display technologies. Nat Biotechnol 29:245–254 . https://doi.org/10.1038/nbt.1791
Deal CE, Balazs AB (2015) Vectored antibody gene delivery for the prevention or treatment of HIV infection. Current Opinion in HIV and AIDS 10:190–197 . https://doi.org/10.1097/coh.0000000000000145
Gąciarz A, Ruddock LW (2017) Complementarity determining regions and frameworks contribute to the disulfide bond independent folding of intrinsically stable scFv. PLoS ONE 12:e0189964 . https://doi.org/10.1371/journal.pone.0189964
Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R (1988) Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proceedings of the National Academy of Sciences 85:5879–5883 . https://doi.org/10.1073/pnas.85.16.5879
Leavy O (2016) The birth of monoclonal antibodies. Nat Immunol 17:S13–S13 . https://doi.org/10.1038/ni.3608
Monnier P, Vigouroux R, Tassew N (2013) In Vivo Applications of Single Chain Fv (Variable Domain) (scFv) Fragments. Antibodies 2:193–208 . https://doi.org/10.3390/antib2020193
Khantasup K, Chantima W, Sangma C, Poomputsa K, Dharakul T (2015) Design and Generation of Humanized Single-chain Fv Derived from Mouse Hybridoma for Potential Targeting Application. Monoclonal Antibodies in Immunodiagnosis and Immunotherapy 34:404–417 . https://doi.org/10.1089/mab.2015.0036
Proba K, Wörn A, Honegger A, Plückthun A (1998) Antibody scFv fragments without disulfide bonds, made by molecular evolution 1 1Edited by I. A. Wilson. Journal of Molecular Biology 275:245–253 . https://doi.org/10.1006/jmbi.1997.1457
Sandomenico A, Sivaccumar JP, Ruvo M (2020) Evolution of Escherichia coli Expression System in Producing Antibody Recombinant Fragments. IJMS 21:6324 . https://doi.org/10.3390/ijms21176324
Additional resources on the Addgene blog:
- Read about frankenbodies, an anti-HA scFv-based probe for fluorescent imaging.
- Learn more about antibodies
- Read about another reagent sharing platform for antibodies, the Developmental Studies Hybridoma Bank
- Learn more about the SunTag system which uses an scFv to amplify the fluorescence intensity of a tagged protein.
Resources on Addgene.org:
- Browse Addgene’s Antibody Plasmid Collection
- Find antibodies for neuroscience research from the Trimmer lab
- Check out Addgene's latest project to develop an open antibody resource
Topics: Antibodies







Leave a Comment